Figure 1.
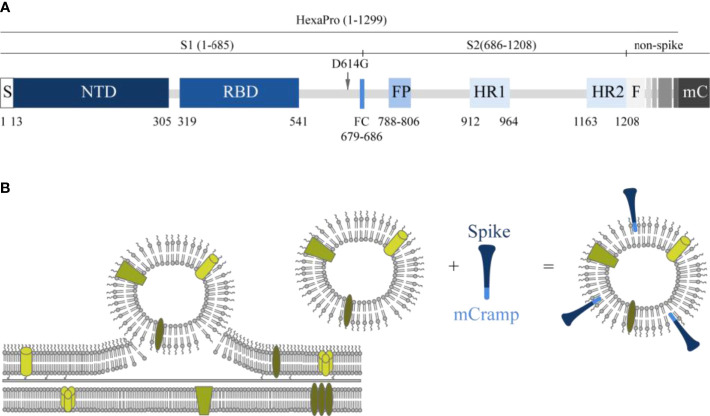
Schematic presentation of the spike molecule and OMV decoration with spike. (A) Schematic of 2019-nCoV S primary structure colored by domain. S, signal sequence; NTD, N-terminal domain; RBD, receptor-binding domain; FP, fusion peptide; HR1–HR2, heptad repeat 1 and 2; FC, disrupted S1/S2 furin cleavage site (R682G, R684S, R685S). Features that were added to the ectodomain (amino acids 1–1,208) expression construct are colored white gray gradient (amino acid 1,209–1,333). From light to dark gray: Foldon trimerization motif (F); HRV 3C site; 8xHis tag; Twin Strep tag; 3x GGGS repeat; mCramp (mC). Not shown: six stabilizing prolines in S2 at positions F817P, A892P, A899P, A942P, K986P, and V987P (27). The aspartic acid at position 614 in the original HexaPro (27) spike protein was replaced with a glycine (D614G). (B) Schematic overview of how antigens can associate with the OMVs. In the spike protein, an mCRAMP (antimicrobial peptide) motif was included which is depicted in light blue. This peptide associates spontaneously to the LPS of the OMV thereby attaching the spike protein (dark blue) to the OMV.