Abstract
Background
The resources of wild ginseng have been reducing sharply, and it is mainly dependent on artificial cultivation in China, Korea and Japan. Based on cultivation modes, cultivated ginseng include understory wild ginseng (the seeds or seedlings of cultivated ginseng were planted under the theropencedrymion without human intervention) and farmland cultivated ginseng (grown in farmland with human intervention). Cultivated ginseng, can only be planted on the same plot of land consecutively for several years owing to soilborne diseases, which is mainly because of the variation in the soil microbial community. In contrast, wild ginseng can grow for hundreds of years. However, the knowledge of rhizosphere microbe communities of the wild ginseng is limited.
Result
In the present study, the microbial communities in rhizosphere soils of the three types of ginseng were analyzed by high-throughput sequencing of 16 S rRNA for bacteria and internal transcribed spacer (ITS) region for fungi. In total, 4,381 bacterial operational taxonomic units (OTUs) and 2,679 fungal OTUs were identified in rhizosphere soils of the three types of ginseng. Among them, the shared bacterial OTUs was more than fungal OTUs by the three types of ginseng, revealing fungal communities were to be more affected than bacterial communities. In addition, the composition of rhizosphere microbial communities and bacterial diversity were similar between understory wild ginseng and wild ginseng. However, higher bacterial diversity and lower fungal diversity were found in rhizosphere soils of wild ginseng compared with farmland cultivated ginseng. Furthermore, the relative abundance of Chloroflexi, Fusarium and Alternaria were higher in farmland cultivated ginseng compared to wild ginseng and understory wild ginseng.
Conclusions
Our results showed that composition and diversity of rhizosphere microbial communities were significantly different in three types of ginseng. This study extended the knowledge pedigree of the microbial diversity populating rhizospheres, and provided insights into resolving the limiting bottleneck on the sustainable development of P. ginseng crops, and even the other crops of Panax.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-021-02421-w.
Keywords: bacteria, fungi, wild ginseng, rhizosphere, soil, Panax ginseng
Background
The rhizosphere is an active area of soil where plant roots and microorganisms interact, and is of great importance for plant health, development, productivity as well as for nutrient cycling [1, 2]. The microbiome can be beneficial or harmful to the host plants. Beneficial microorganisms provide nutrients from the soil and protect the plant from pathogens, enhancing tolerance to abiotic stresses [3]. However, pathogenic microorganisms slow down plant growth, reduce survival rate, and cause yield loss [4]. In recent years, many studies have been conducted to characterize rhizosphere microbiome in plant varieties, such as barley, wheat, soybean and blueberry [5–8]. These studies mainly showed that plant varieties can affect the rhizosphere microbial diversity and composition. Therefore, a better understanding the effects of the composition and function in the rhizosphere microbial community will contribute to plant breeding and open up a new approach for the rational use of plant-microbial interactions in agriculture.
The ginseng (Panax ginseng C. A. Meyer.) is used as a traditional Chinese medicine to treat many diseases, due to its anti-inflammatory and antitumor compounds [9]. Ginseng belongs to the family Araliaceae that is distributed in Asia, particularly in Korea and China [10]. Wild ginseng germplasm resources are scarce due to excessive land exploitation and disruption of the environment; thus, wild ginseng has been gradually replaced by cultivated ginseng in the market [11]. There are two kinds of cultivated ginseng, farmland cultivated ginseng and understory wild ginseng. Farmland cultivated ginseng is planted in farmland that was once forested with agricultural management, such as artificial shading and spraying pesticides, and farmland cultivated ginseng grows very quickly. In contrast, understory wild ginseng refers to cultivated ginseng seeds or seedlings grown under natural forest conditions for many years with little human interference. Its morphology and intrinsic quality characteristics of roots are similar to those of wild ginseng [12].
Cultivated ginseng, especially farmland cultivated ginseng, is susceptible to various soil-borne diseases. Farmland cultivated ginseng roots are harvested 5–6 years after planting, and the survival rate of farmland cultivated ginseng seedlings was less than 25% after 3 years [13]. Previous studies have showed Chloroflexi and Fusarium can cause root rot and root rust, and that these are the two most common soil-borne diseases, resulting in reduced ginseng quality and production [14–16]. In addition, root rot and root rust are also catastrophic diseases of other crops of Panax, like Panax quinquefolium and P. notoginseng [17, 18]. So bacterial and fungal community in rhizosphere soils changes during cultivation of these crops had received much attention [19]. It has been shown that microbial community in rhizosphere soil of ginseng is affected by cultivation ages, developmental stages and cultivation modes [13, 20–22]. Dong et al. (2017) found that fungal diversity increased, whereas bacterial diversity decreased in the rhizosphere soils at the root growth stage of ginseng [22]. Dong et al. (2018) also found that soil bacterial diversity decreased with the increase of ginseng planting years by comparing 1, 2 and 3 year old ginseng [21].
However, wild ginseng can grow in natural environments for decades or even hundreds of years [23]. Recent findings suggested that, one of the roles of rhizosphere microorganisms is to protect plants from pathogen infection by promoting the cascade modification of beneficial microorganism [24, 25]. These raise the question: the root microorganisms of wild ginseng are different from cultivated ginseng, and can this difference protect wild ginseng from pathogenic microbes? Whereas the structure in the rhizosphere microbial communities of wild ginseng is not clear. Therefore, in this study, we explored the diversity and structure of the bacterial and fungal communities in rhizosphere soil of wild ginseng, and also compared the rhizosphere microbial communities of wild ginseng and two types of cultivated ginseng. The study will provide insights into underlying mechanisms of ginseng planting and disease resistance.
Results
Rhizosphere community diversity in three types of ginseng
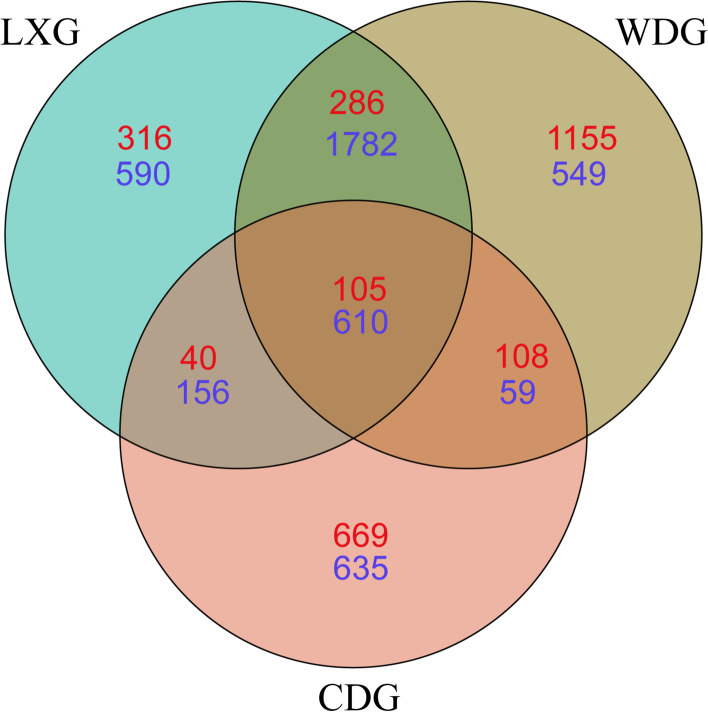
To explore the bacterial and fungal communities in the rhizosphere soil of understory wild ginseng (LXG), farmland cultivated ginseng (CDG) and wild ginseng (WDG), we obtained 1,135,354 high-quality paired reads by high-throughput sequencing of 16S rRNA for bacteria and 797,696 paired reads by high-throughput sequencing of internal transcribed spacer (ITS) region for fungi. In total, 3,138 bacterial operational taxonomic units (OTUs) and 747 fungal OTUs were identified in LXG, 1,460 bacterial OTUs and 922 fungal OTUs were identified in CDG, while 3,000 bacterial and 1,654 fungal OTUs were identified in WDG, respectively (Fig. 1). The percentage of shared OTUs between LXG and WDG was smaller for the fungal community than for the bacterial community, and this pattern also occurred between LXG and CDG and between WDG and CDG (Fig. 1).
Fig. 1.
Venn diagrams of shared bacterial (blue) and fungal (red) OTUs in rhizosphere of three types of ginseng. LXG, understory wild ginseng; CDG, farmland cultivated ginseng; WDG, wild ginseng
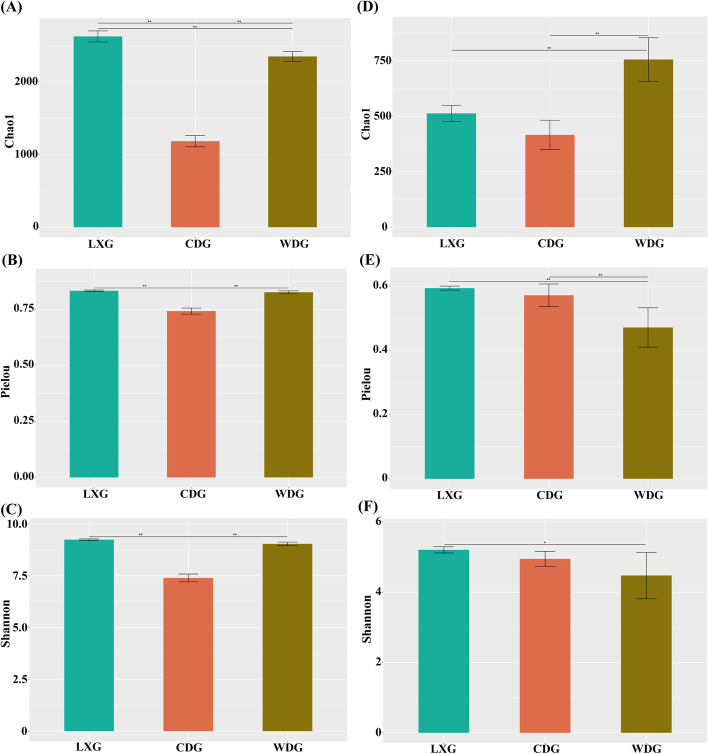
The rhizosphere microbial diversity differed among three types of ginseng (Fig. 2). The bacterial alpha diversity of LXG and WDG were similar, however, that of CDG was significantly lower than those of WDG (p < 0.01) (Fig. 2 A, B, C). WDG had the highest fungal species richness (Chao 1), however, the fungal species evenness (Pielou) and fungal species diversity (Shannon) of WDG was the lowest (p < 0.01) (Fig. 2D, E, F). The fungal alpha diversity of LXG was higher than that of CDG (Fig. 2D, E, F).
Fig. 2.
Chao 1, Pielou and Simpson indexes in the bacteria (A), (B), (C) and fungi (D), (E), (F) from three types of ginseng in rhizosphere, data were means ± standard error. LXG, understory wild ginseng; CDG, farmland cultivated ginseng; WDG, wild ginseng. *Significant at the 0.05 probability level. **Significant at the 0.01 probability level
Bacterial and fungal community composition
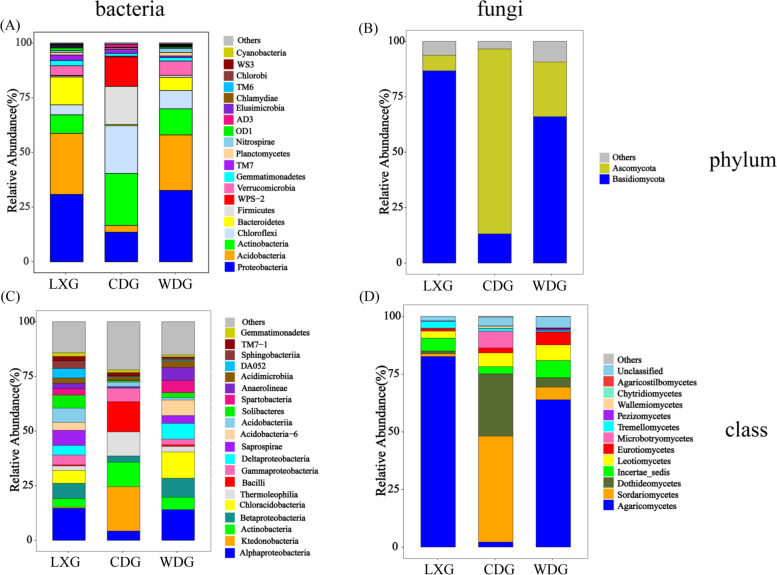
The composition and abundance for each taxon were obtained based on the OTU classification results. For bacteria, the dominant phyla were Proteobacteria, Acidobacteria, Actinobacteria and Chloroflexi in LXG (relative abundances of 30.76%, 27.92%, 8.51% and 4.57%, respectively) and WDG (relative abundances of 32.61%, 25.39%, 11.92% and 8.39%, respectively) (Fig. 3 A), However, Actinobacteria (23.75%) was the most phylum in CDG, followed by Chloroflexi (21.85%), Firmicutes (17.40%) and Proteobacteria (13.53%). The ANOVA analysis suggested that the proportions of each main four phyla were not significantly different between LXG and WDG, however, these four phyla (Proteobacteria, Acidobacteria, Actinobacteria and Chloroflexi) were significantly different between WDG and CDG, LXG and CDG, respectively (Additional files 2: Fig. S2, p < 0.01). The most abundant bacterial classes were Alphaproteobacteria and Betaproteobacteria in LXG (relative abundances of 14.71% and 7.07%, respectively) and WDG (relative abundances of 14.02% and 8.76%, respectively), then Saprospirae (6.94%) and Acidobacteria (6.94%) in LXG, after Chloracidobacteria (12.02%) and Deltaproteobacteria (7.17%) in WDG. Ktedonobacteria (20.24%), Bacilli (13.80%), Actinobacteria (11.18%) and Thermoleophilia (11.05%) had the highest relative abundances in CDG (Fig. 3 C).
Fig. 3.
The composition of bacterial and fungal community from different types of ginseng rhizosphere. The phylum level of bacteria (A) and fungi (B), and the class level of bacteria (C) and fungi (D). The relative abundances in the top 20 were chosen to exhibit. Others represented of low relative abundance that ranks lower than top 20. LXG, understory wild ginseng; CDG, farmland cultivated ginseng; WDG, wild ginseng
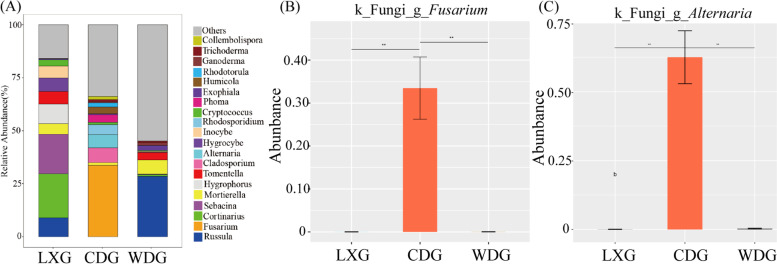
For fungi, Ascomycota and Basidiomycota accounted for more than 90% of the total abundance across LXG, CDG and WDG (Fig. 3B), but the result of ANOVA indicated that the proportions of each phylum were different in each type of ginseng (Additional files 2: Fig. S3, p < 0.01). At the class level, the dominant classes were Agaricomycetes, incertae_sedis and Leotiomycetes in LXG (relative abundances of 82.56%, 5.61% and 3.04%, respectively) and WDG (relative abundances of 63.85%, 7.45% and 6.78%, respectively) (Fig. 3D). In CDG, the relative abundance of Sordariomycetes (45.92%) was the highest, followed by Dothideomycetes (27.00%) and Microbotryomycetes (7.22%) (Fig. 3D). At the genus level, the most abundant genus was Fusarium (28.37%) in CDG; however, it was rare in LXG (< 0.01%) and WDG (0.01%) (Fig. 4 A), and ANOVA suggested that the abundance of Fusarium was the highest in CDG (p < 0.01) (Fig. 4B). In addition, the abundance of Alternaria in CDG was higher than in LXG and WDG (p < 0.01) (Fig. 4 C).
Fig. 4.
The composition of fungal community at the genus level (A). The relative abundances in the top 20 were chosen to exhibit. Others represented of low relative abundance that ranks lower than top 20. The relative abundance of Fusarium (B) and Alternaria (C) in the rhizosphere for three types of ginseng, data were means ± standard error. LXG, understory wild ginseng; CDG, farmland cultivated ginseng; WDG, wild ginseng. *Significant at the 0.05 probability level. **Significant at the 0.01 probability level
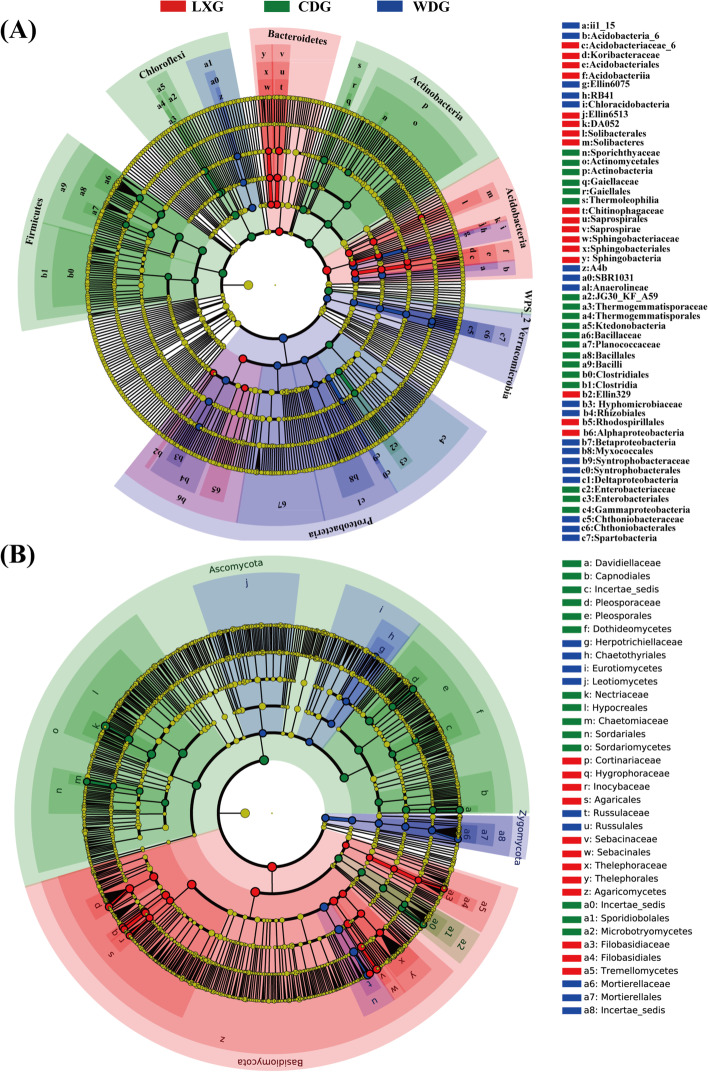
The LEfSe analysis of the rhizosphere bacterial communities showed that there were 68 differentially abundant taxa among the three types of ginseng. Of the 68 taxa, 23 were differentially abundant in WDG (Fig. 5 A, Additional files 2: Fig. S4A), namely the Verrucomicrobia phylum and the Deltaproteobacteria, Betaproteobacteria, Acidobacteris_6, Chloracidobacteria and Anaerolineae classes. The enriched taxa in LXG were the phyla Bacteroidetes and Acidobacteria and the class Alphaproteobacteria. The differentially abundant taxa in the rhizosphere soils of CDG were the Firmicutes, Actinobacteria, WPS-2 and Chloroflexi phyla and the Gammaproteobacteria class.
Fig. 5.
LEfSe analysis showing the different rhizosphere taxa among three types of ginseng in bacteria (A) and fungi (B). The diameter of each circle is proportional to the relative abundance of the taxon. The inner to outer circle corresponds to the level of the phylum to the genus. LXG, understory wild ginseng; CDG, farmland cultivated ginseng; WDG, wild ginseng
The LEfSe analysis of the fungal communities from LXG, CDG and WDG showed that, there were 69 differentially abundant taxa with an LDA score higher than 2.0 (Fig. 5B, Additional files 2: Fig. S4B). Among the 69 fungal taxa, 15 fungal taxa were differentially abundant in WDG, principally including the Mortierellomycota phylum, the Leotiomycetes and Eurotiomycetes classes, and the Russulales order. The abundant taxa in the rhizosphere soils of LXG were the Agaricomycetes and Tremellomycetes classes. The most differentially abundant fungal taxa were the Sordariomycetes, Dothideomycetes and Microbotryomycetes classes in CDG.
Factors driving rhizosphere microbial communities in three types of ginsengs
Effects of soil physical and chemical properties and plant types on the structure of microbial communities in the rhizosphere of ginseng were analyzed using 4, 381 bacterial OTUs and 2, 679 fungal OTUs. The result of mantel test showed that bacterial and fungal community were correlated with soil physical and chemical properties, but not significant (Table 1, p > 0.05). However, the PERMANOVAs results suggested ginseng type explained 90.118% and 84.699% of variance in bacterial and fungal communities, respectively (Table 2, p < 0.01). The PCoA of Bray-Curtis distance matrix demonstrated that samples of LXG, CDG and WDG showed clear separation, suggesting that the bacterial and fungal communities were obviously different among three types of ginseng (Additional files 2: Fig. S1).
Table 1.
Mantel tests of the influence of soil physical and chemical properties on microbial communities associated
| bacteria | fungi | |||
|---|---|---|---|---|
| Soil parameter | R2 | p | R2 | p |
| TP | -0.45 | 0.67 | -0.93 | 1.00 |
| TN | -0.45 | 0.67 | -0.93 | 1.00 |
| TK | -0.45 | 0.67 | -0.93 | 1.00 |
| pH | 1.00 | 0.34 | 0.78 | 0.33 |
| Sand | 0.89 | 0.17 | 0.36 | 0.50 |
| Silt | 0.89 | 0.17 | 0.35 | 0.50 |
| Clay | -0.99 | 1.00 | -0.63 | 0.83 |
Note: TP, total phosphorus; TN, total nitrogen; TK, total potassium; Sand, the content of sand in soil; Silt, the content of silt in soil; Clay, the content of clay in soil; R2, correlation coefficient; p, p-value
Table 2.
PERMANOVAs of the influence of ginseng types on microbial communities associated
| bacterial community | fungal community | |||||
|---|---|---|---|---|---|---|
| factor | F | R2 | p | F | R2 | p |
| types | 68.3980 | 0.90118 | < 0.01 | 41.5160 | 0.84699 | < 0.01 |
Note: F, F.model; R2, Variation ; p, p-value
Discussion
The factor affected the rhizosphere microbial community
Owing to the lack of wild ginseng germplasm resources and the low survival rate of cultivated ginseng, it is very difficult to collect the rhizosphere soils of cultivated ginseng and wild ginseng in the same field. This phenomenon is common among wild species. Wang et al. (2018) compared the rhizosphere bacterial diversity of four Ferulic species from four locations based on 16S rRNA sequencing [26]. Nevertheless, the three sampling locations of our study were all in Jilin Province of China. In addition, we also downloaded the data of soil physical and chemical properties to explore the influence of soil physical and chemical properties on microbial communities. The mantel test suggested soil physical and chemical properties effected bacterial and fungal communities, but not significant. However, PERMANOVAs result indicated that the ginseng types significantly affected the rhizosphere microbial community. Christine et al. (2007) found that there was no difference in soil microbial characteristics between organic soil and conventional fertilized soil, and the type of plant has a great influence on microbial biomass [27]. Muhammad et al. (2020) also suggested plant types had stronger effects on soil microbial communities than biochar or fertilizer [28]. Therefore, we considered that the effect of soil physical and chemical properties on bacterial and fungal communities is not obvious. The type of ginseng might have a great influence on the rhizosphere microbial community.
In our study, we found that the proportion of shared fungal OTUs was much smaller than that of bacterial OTUs between any two pairs of LXG, CDG and WDG. We inferred fungal communities appeared to be more affected than bacterial communities. Coleman-Derr et al. (2016) suggested that fungal communities were perhaps more shaped by geographic distance than bacterial communities in rhizosphere soils of Agave from California and Mexico [29]. Wang et al. (2021) also found the fungal communities were more influenced than the bacterial communities by different ginseng cultivars [30]. These studies showed that fungal communities are more affected than bacterial communities, which is consistent with our results.
Different bacterial diversity and fungal diversity in three types of ginsengs
The bacterial alpha diversity of LXG and WDG were similar, and WDG shared more bacterial OTUs with LXG than with CDG. Maybe the understory wild ginseng and wild ginseng both grow in the theropencedrymion, moreover, understory wild ginseng is a semi-wild ginseng and its morphology and intrinsic quality characteristics of roots are similar to those of wild ginseng [12, 31]. However, the bacterial alpha diversity of CDG decreased compared with that of WDG, showing a loss of natural bacterial diversity in the rhizosphere. A number of agronomic management practices potentially influenced bacterial diversity in farmland cultivated ginseng. Ginseng is a shade-loving plant, so farmland cultivated ginseng need to be artificially shaded and irrigated during the growth process [32]. These treatments effected the soil moisture and regional temperature of cultivated ginseng. Nuccio et al. (2016) compared rhizosphere microorganisms from three California prairie wild oats (Avena spp.), suggesting rhizosphere microorganisms were influenced by factors related to the regional climate (soil moisture and temperature) [33]. Therefore, artificial shading and irrigation treatment affected the bacterial diversity. In addition, farmland cultivated ginseng was susceptible to diseases during cultivation, so pesticides were sprayed to reduce the impact of pests and diseases [32, 34]. Pesticides might have indirectly affected root exudates or directly inhibited the reproduction of certain rhizosphere microorganisms during ginseng cultivation [35, 36]. Thus, pesticide disposal also reduced the microbial diversity of farmland cultivated ginseng. Furthermore, the decrease in bacterial diversity might also be associated to the host plants. The rhizosphere microbial diversity of domesticated sugar beets decreased compared to their wild ancestors [37]. Cultivated crops had the characteristics of fast growth and high yield, which might lead to different amounts and types of organic compounds secreted from the roots, resulting in different subsurface microbial community structures [38].
In contrast, fungal diversity showed the opposite phenomenon. We found that the fungal diversity (Shannon and Pielou) in CDG was higher than that in WDG. But the Chao 1 was the highest in WDG, which may be associated with the greater number of fungal OTUs in WDG than in CDG and LXG. A similar phenomenon has been observed in soybean and their wild species, and the fungal diversity of cultivated soybean increased compared to its wild type [39]. Selective breeding of modern crops perhaps had promoted the proliferation of specific crop-related microbial taxa, leading to increased fungal diversity in cultivated ginseng [40]. Furthermore, the fungal alpha diversity of LXG was higher than CDG, which might be related to high content of ginsenoside for understory wild ginseng. According to previous studies, the fungal diversity of soil was affected by the content of ginsenosides in ginseng growth [41, 42]. Yong et al. (2007) also found understory wild ginseng contained higher amounts of ginsenosides than farmland cultivated ginseng [43]. Therefore, understory wild ginseng secreted higher ginsenosides to the rhizosphere soil, resulting in higher diversity of rhizosphere fungi.
Changes in the composition of microbial communities among the three types of ginsengs
In the composition of bacterial communities, our study indicated that Proteobacteria and Acidobacteria all existed in LXG, CDG and WDG, and previous studies also confirmed that Proteobacteria and Acidobacteria were the dominant populations in the rhizosphere soil of ginseng [44]. The relative abundance of Chloroflexi in CDG was higher than that in WDG and LXG (p < 0.01). Wang et al. (2019) suggested that root rust may be caused by Chloroflexi in rhizosphere microbial communities based on five cultivated ginseng samples with different severity of rusty root disease [15]. Although the rhizosphere soils in our study came from healthy ginseng, Chloroflexi was a hidden danger that causes the disease of farmland cultivated ginseng. In addition, Verrucomicrobia was significantly high abundance in WDG and also has been found in the rhizosphere of the common bean, and Verrucomicrobia was also mainly found in wild bean accessions [38]. Verrucomicrobia probably had established beneficial links with wild species to protect wild species from pathogens. However, there are few studies on Verrucomicrobia, which still need further research.
In fungi community, the same fungal phyla were detected but the fungal community composition, also at phylum level, the relatively abundances of Basidiomycota and Ascomycota was different in LXG, CDG and WDG. Ascomycota and Basidiomycota also were the dominant phyla in the rhizosphere soil of P. notoginseng [45]. Ascomycota, which has an important role in the decomposition of soil organic matter and largely dominates the active fungal community through the assimilation of root exudates [46]. Furthermore, the main pathogenic fungus Fusarium that caused root rot belongs to the Ascomycota, which was the most predominant phylum in CDG. Fusarium was a potential phytopathogen (includes potential pathogens) that can cause root rot in various species, including ginseng, American ginseng, soybean and sunflower [47–50]. We found that the abundance of Fusarium was the highest in CDG, while that in WDG was only 0.01% (p < 0.01). Likewise, cultivated rice had a higher abundance of pathogens comparing with the wild varieties [39]. Moreover, Alternaria was a pathogenic fungi associated with ginseng rusty roots [51]. And the abundance of Alternaria in CDG was higher than that of LXG and WDG. This further suggests that domesticated crops reduced their ability to establish beneficial associations with the rhizosphere microbiome, thus stimulating the spread of pathogens in rhizosphere [38]. This also explains how wild ginseng can grow in natural environments for decades or even hundreds of years. In addition, continuous planting may also lead to an increase in rhizosphere pathogens. The pathogens, including Alternaria and Fusarium, that were highly enriched in the rhizosphere of 30-year sugar beet [52]. This result provided new scientific insights for the healthy growth, reducing the incidence of ginseng and improving the yield of ginseng. Through a comprehensive understanding of wild ginseng rhizosphere microorganisms, farmland soil can be improved to create a suitable soil environment for ginseng growth and reduce forest logging during ginseng planting.
Conclusions
In this study, we systematically studied the rhizosphere microbial communities of three types of ginseng, including LXG, CDG and WDG. The results showed soil physical and chemical properties effected bacterial and fungal communities, but not significantly. However, the type of ginseng had a great influence on rhizosphere microbial communities. We found fungal communities were more susceptible than bacterial communities. By comparing rhizosphere microbe of three types of ginseng, the composition of rhizosphere microbial communities in LXG and in WDG was similar. There were significant differences in the composition of rhizosphere microbial community in WDG and in CDG. In addition, the bacterial diversity of WDG and LXG was also similar. The higher bacterial diversity and lower fungal diversity in WDG compared with CDG. Furthermore, the relative abundance of Chloroflexi, Fusarium and Alternaria were higher in CDG compared to WDG and LXG. This result may provide insights for ginseng breeding and yield improvement, and supply feasible information for soil management.
Methods
Sampling Sites and Samples Collection
Rhizosphere soil samples were collected from three type of ginsengs, including understory wild ginseng (LXG), farmland cultivated ginseng (CDG) and wild ginseng (WDG). Rhizosphere soil samples in this study were collected in August 2018. A collection of ginseng rhizosphere soil was made, and samples were taken from a depth of 20 cm using a sterile shovel. Ginseng plants were carefully removed from the ground, keeping the root system intact. The large clumps of soil on the roots were removed, then brushed soil attached to the roots with a brush. Each soil samples were passed through a 2 mm sieve, finally into a sterile tube. The soil samples in each type are from at least three healthy, disease-free roots of ginseng (one to three rhizosphere soil samples were collected from the roots of each ginseng). In total, the rhizosphere soils samples of LXG, CDG and WDG were set up with seven, five and six, respectively (Additional files 1: Table S1). All samples were then transported to the liquid nitrogen within one hour and immediately transported to the laboratory. Finally, the soil samples were stored at −80 °C until genomic DNA extraction using an E.Z.N.A.® Stool DNA Kit (Omega, Shanghai).
All ginsengs were grown for about 15 years. wild ginseng and understory wild ginseng both grow in theropencedrymion, and there were similar vegetation types under the forest. The rhizosphere samples of understory wild ginseng were collected from Linjiang city of Jilin Province, and the soil was Mollic Albi-boric Cambosols (sand 51%, silt 32%, clay 17%), then chemical characteristics of the soil were (mg kg−1): 82 (P), 199 (N) and 2198 (K) with pH (5.99). The soil samples of farmland cultivated ginseng were from Ji’an city of Jilin Province, and the soil was Mollic bori-Udic Cambosols (sand 33%, silt 49%, clay 18%), and chemical characteristics of the soil were (mg kg−1): 146 (P), 401 (N) and 1823 (K) with pH (6.62). The rhizosphere soils of wild ginseng were collected from Korean autonomous county, Jilin Province. The soil was Mollic bori-Udic Cambosols (sand 45%, silt 37%, clay 18%), and chemical characteristics of the soil were (mg kg−1): 146 (P), 401(N) and 1823 (K) with pH (5.99) (Table 3). The soil characteries were mapped using the National Earth System Science Data Center [53].
Table 3.
The physical and chemical properties of the soil in sampling locations for ginseng
| LXG | CDG | WDG | |
|---|---|---|---|
| soil taxonomy | Mollic Albi-boric Argosolos | Mollic bori-Udic Cambosols | Mollic bori-Udic Cambosols |
| TP (mg kg−1) | 82 | 146 | 146 |
| TN (mg kg−1) | 199 | 401 | 401 |
| TK (mg kg−1) | 2198 | 1823 | 1823 |
| pH | 5.99 | 6.62 | 5.99 |
| Sand (%) | 51 | 33 | 45 |
| Silt (%) | 32 | 49 | 37 |
| Clay (%) | 17 | 18 | 18 |
Note: TP, total phosphorus; TN, total nitrogen; TK, total potassium; Sand, the content of sand in soil; Silt, the content of silt in soil; Clay, the content of clay in soil. LXG, understory wild ginseng; CDG, farmland cultivated ginseng; WDG, wild ginseng
PCR, amplicon quantification, HiSeq library construction and sequencing
The variable V3-V4 region of the bacterial 16S rRNA gene and fungal ITS1 region were amplified from each sample with the primers pairs 341F (5’-ACTCCTACGGGAGGCAGCAG-3’) / 806R (5’-GGACTACHVGGGTWTCTAAT-3’) and ITS-1 (5’-CTTGGTCATTTAGAGGAAGTAA-3’)/ITS-2 (5’-GCTGCGTTCTTCATCGATGC-3’) [54, 55]. All PCRs reactions were performed using NEB Phusion High-Fidelity PCR Master Mix following the manufacturer’s recommendations. The PCR reaction system contained 30 ng of DNA, 4µL of PCR primer mix and 25µL of PCR Master Mix. The following PCR conditions were used: 98 °C for 3 min; followed by 30 cycles of 98 °C for 45 s, 55 °C for 45 s and 72 °C for 45 s; and a final extension of 72 °C for 7 min. Then, the PCR products integrity was subjected to 1% agarose gel electrophoresis and purified using Ampure XP beads (Beckman, America) to remove the unspecific products. The final library was quantitated in two ways: determination of the average molecule length using an Agilent 2100 bioanalyzer instrument (Agilent DNA 1000 reagents, America), and quantification of the library by real-time quantitative PCR. The qualified libraries were sequenced pair-end on the system with the sequencing strategy PE250 under the HiSeq platform (Illumina, America).
Data analysis and statistics
After removing reads with ambiguous bases, an average Phred score less than 20 and the length lower than 10 bp, the remaining reads were merged into Tags based on overlapping regions using FLASH (fast length adjustment of short reads, v1.2.11) within paired-end reads [56]. Then, operational taxonomic units (OTUs) were clustered with a 97% similarity cut off by using UPARSE (v7.0.1090) [57]. The chimeric sequences were identified and removed using UCHIME software (v4.2.40) [58]. OTUs undoubtedly belonging to chloroplasts or mitochondria were also removed. Subsequently, the taxonomic classification of the representative sequence for each bacterial and fungal OTUs was annotated using Greengenes (v201304) and UNITE (v7.2) reference databases, respectively, with the RDP Classifier (Ribosomal Database Project, v2.2). The taxa with significant different in rhizosphere microbial community among three type of ginsengs was performed by ANOVA (One-way Analysis of Variance, p < 0.05) in SPSS_Statistics_23. A venn plotter was used to obtain the number of unique and common OTUs using the ‘VennDiagram’ R package (v3.1.1). The alpha diversity of the bacterial and fungal communities was calculated with the Chao 1 (species richness), Pielou (species evenness) and Shannon (species diversity) indexes for each sample using in MOTHUR (v1.31.2) [59]. The differences in alpha diversity among the three types of ginseng were determined by Tukey’s honestly significant difference test in R package (v3.1.1) (p < 0.05). Mantel tests and Permutational ANOVAS (PERMANOVAs) were performed to assess the correlation between rhizosphere microbial communities and soil physical and chemical properties, type of ginseng by using R package ‘vegan’ (v2.5-7), respectively [60]. Principal coordinate analysis (PCoA) was performed in QIIME software (v1.80) to reflect the beta diversity of the microbial community, evaluate the similarity in community composition among the different types of ginseng based on the Bray-Curtis distance matrix [61]. Linear discriminant analysis (LDA) effect sizes (LEfSe) was used to detect notably different taxa among the samples using the Galaxy online analytics platform, and LEfSe identity different abundant taxa with an linear discriminant analysis (LDA) score higher than 2.0 (http://huttenhower.sph.havard.edu/galaxy).
Supplementary Information
Acknowledgements
We thank reviewers and editors for their constructive comments that greatly improved our manuscript. We also thank Prof. Hongxing Xiao and Wei Zhang for helping to collected the samples in this study.
Authors' contributions
M. S. and X. F. designed the experiments. X. F. and H. W. performed most of experiments and analyzed the data. Other authors assisted in experiments and discussed the results. X. F. and H. W. wrote the manuscript. The author(s) read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (grant number: 31770243).
Availability of data and materials
The sequencing dataset analyzed during the current study is available in the NCBI Sequence Read Archive (PRJNA701796 and PRJNA701800).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhalnina K, Louie KB, Hao Z, Mansoori N, Da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nature Microbiology. 2018;3(4):470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson D, Watson CA. The beneficial rhizosphere: A dynamic entity. Applied Soil Ecology. 2000;15(2):99–104. [Google Scholar]
- 3.Jacoby R, Peukert M, Succurro A, Koprivova A, Kopriva S: The role of soil microorganisms in plant mineral nutrition—current knowledge and future directions. Front Plant Sci. 2017;8:1617. [DOI] [PMC free article] [PubMed]
- 4.Yin C, Casa Vargas JM, Schlatter DC, Hagerty CH, Hulbert SH, Paulitz TC. Rhizosphere community selection reveals bacteria associated with reduced root disease. Microbiome. 2021;9(1):86. [DOI] [PMC free article] [PubMed]
- 5.Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host and Microbe. 2015;17(3):392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahoney AK, Yin C, Hulbert SH. Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars. Front Plant Sci. 2017;8:132. [DOI] [PMC free article] [PubMed]
- 7.Zhong Y, Yang Y, Liu P, Xu R, Rensing C, Fu X, Liao H. Genotype and rhizobium inoculation modulate the assembly of soybean rhizobacterial communities. Plant Cell and Environment. 2019;42(6):2028–2044. doi: 10.1111/pce.13519. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Wang W, Shen Z, Wang J, Chen Y, Wang D, Liu G, Han M: Comparison and interpretation of characteristics of Rhizosphere microbiomes of three blueberry varieties. BMC Microbiol 2021, 21(1). [DOI] [PMC free article] [PubMed]
- 9.Ernst E. Panax ginseng: An overview of the clinical evidence. Journal of Ginseng Research. 2010;34(4):259–263. [Google Scholar]
- 10.Baeg IH, So SH. The world ginseng market and the ginseng (Korea) Journal of Ginseng Research. 2013;37(1):1–7. doi: 10.5142/jgr.2013.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Yang HY, You XL, Li YH. Diversity of endophytic fungi from roots of Panax ginseng and their saponin yield capacities. SpringerPlus. 2013;2(1):1–9. doi: 10.1186/2193-1801-2-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun H, Wang Q, Zhang Y, Yang Z, Xu C. Integrated Evaluation of Soil Fertility of Panax ginseng under Different Cultivation Modes. Journal of Jilin Agricultural University. 2015;37(03):323–331. [Google Scholar]
- 13.Ying YX, Ding WL, Li Y. Characterization of soil bacterial communities in rhizospheric and nonrhizospheric soil of panax ginseng. Biochemical Genetics. 2012;50(11-12):848–859. doi: 10.1007/s10528-012-9525-1. [DOI] [PubMed] [Google Scholar]
- 14.Chanyong L, Kim KY, Lee JE, Kim S, Ryu D, Choi JE, An G. Enzymes hydrolyzing structural components and ferrous ion cause rusty-root symptom on ginseng (Panax ginseng) Journal of Microbiology and Biotechnology. 2011;21(2):192–196. doi: 10.4014/jmb.1008.08010. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Sun H, Xu C, Ma L, Li M, Shao C, Guan Y, Liu N, Liu Z, Zhang S, et al. Analysis of rhizosphere bacterial and fungal communities associated with rusty root disease of Panax ginseng. Applied Soil Ecology. 2019;138:245–252. [Google Scholar]
- 16.Miao ZQ, Li SD, Liu X, Chen YJ, Li YH, Wang Y, Guo RJ, Xia ZY, Zhang KQ. The causal microorganisms of Panax notoginseng root rot disease. Scientia Agricultura Sinica. 2006;39:1371–1378. [Google Scholar]
- 17.Ma L, Cao YH, Cheng MH, Huang Y, Mo MH, Wang Y, Yang JZ, Yang FX. Phylogenetic diversity of bacterial endophytes of Panax notoginseng with antagonistic characteristics towards pathogens of root-rot disease complex. Antonie van Leeuwenhoek, International Journal of General and Molecular Microbiology. 2013;103(2):299–312. doi: 10.1007/s10482-012-9810-3. [DOI] [PubMed] [Google Scholar]
- 18.Wu Z, Hao Z, Sun Y, Guo L, Huang L, Zeng Y, Wang Y, Yang L, Chen B. Comparison on the structure and function of the rhizosphere microbial community between healthy and root-rot Panax notoginseng. Applied Soil Ecology. 2016;107:99–107. [Google Scholar]
- 19.Jiang J, Yu M, Hou R, Li L, Ren X, Jiao C, Yang L, Xu H. Changes in the soil microbial community are associated with the occurrence of Panax quinquefolius L. root rot diseases. Plant and Soil. 2019;438(1-2):143–156. [Google Scholar]
- 20.Xiao C, Yang L, Zhang L, Liu C, Han M. Effects of cultivation ages and modes on microbial diversity in the rhizosphere soil of panax ginseng. Journal of Ginseng Research. 2016;40(1):28–37. doi: 10.1016/j.jgr.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong L, Xu J, Li Y, Fang H, Niu W, Li X, Zhang Y, Ding W, Chen S. Manipulation of microbial community in the rhizosphere alleviates the replanting issues in Panax ginseng. Soil Biol Biochem. 2018;125:64–74. [Google Scholar]
- 22.Dong L, Xu J, Zhang L, Cheng R, Wei G, Su H, Yang J, Qian J, Xu R, Chen S. Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharmaceutica Sinica B. 2018;8(2):272–282. doi: 10.1016/j.apsb.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H. A brief discussion on the classification and identification of wild Ginseng. Chinese Rural Medicine. 2021;28(01):13–14. [Google Scholar]
- 24.Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. Interplay between Innate Immunity and the Plant Microbiota. Annual Review of Phytopathology. 2017;55:565–589. doi: 10.1146/annurev-phyto-080516-035623. [DOI] [PubMed] [Google Scholar]
- 25.Schlatter D, Kinkel L, Thomashow L, Weller D, Paulitz T. Disease suppressive soils: New insights from the soil microbiome. Phytopathology. 2017;107(11):1284–1297. doi: 10.1094/PHYTO-03-17-0111-RVW. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Wang Z, Jiang P, He Y, Mu Y, Lv X, Zhuang L: Bacterial diversity and community structure in the rhizosphere of four Ferula species. Scientific Reports 2018, 8(1). [DOI] [PMC free article] [PubMed]
- 27.Stark C, Condron LM, Stewart A, Di HJ, O’Callaghan M. Effects of past and current crop management on soil microbial biomass and activity. Biology and Fertility of Soils. 2007;43(5):531–540. [Google Scholar]
- 28.Azeem M, Sun D, Crowley D, Hayat R, Hussain Q, Ali A, Tahir MI, Jeyasundar PGSA, Rinklebe J, Zhang Z. Crop types have stronger effects on soil microbial communities and functionalities than biochar or fertilizer during two cycles of legume-cereal rotations of dry land. Sci Total Environ. 2020;715:136958. [DOI] [PubMed]
- 29.Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martinez LP, Tringe SG. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytologist. 2016;209(2):798–811. doi: 10.1111/nph.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Fang X, Wu H, Cai X, Xiao H. Effects of plant cultivars on the structure of bacterial and fungal communities associated with ginseng. Plant and Soil. 2021;465(1-2):143–156. [Google Scholar]
- 31.Li L, Qu Y, Dai W, S, Ma Q, K, Fang H, Y. Comparative research progress of chemical constituents of Panax ginseng in different habitats. Journal of Guangdong Pharmaceutical University. 2018;34(06):803–807. [Google Scholar]
- 32.Huang X. Study on cultivation management and pest control of ginseng in field. Agriculture and Technology. 2019;39(02):119–120. [Google Scholar]
- 33.Nuccio EE, Anderson-Furgeson J, Estera KY, Pett-Ridge J, De Valpine P, Brodie EL, Firestone MK. Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology. 2016;97(5):1307–1318. doi: 10.1890/15-0882.1. [DOI] [PubMed] [Google Scholar]
- 34.Rahman M, Punja ZK. Influence of iron on cylindrocarpon root rot development on ginseng. Phytopathology. 2006;96(11):1179–1187. doi: 10.1094/PHYTO-96-1179. [DOI] [PubMed] [Google Scholar]
- 35.Chen S, Li X, Lavoie M, Jin Y, Xu J, Fu Z, Qian H. Diclofop-methyl affects microbial rhizosphere community and induces systemic acquired resistance in rice. Journal of Environmental Sciences (China) 2017;51:352–360. doi: 10.1016/j.jes.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Arango L, Buddrus-Schiemann K, Opelt K, Lueders T, Haesler F, Schmid M, Ernst D, Hartmann A. Effects of glyphosate on the bacterial community associated with roots of transgenic Roundup Ready® soybean. European Journal of Soil Biology. 2014;63:41–48. [Google Scholar]
- 37.Zachow C, Müller H, Tilcher R, Berg G. Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima-ancestor of all beet crops-and modern sugar beets. Front Microbiol. 2014;5:415. [DOI] [PMC free article] [PubMed]
- 38.Pérez-Jaramillo JE, Mendes R, Raaijmakers JM. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Molecular Biology. 2016;90(6):635–644. doi: 10.1007/s11103-015-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi S, Chang J, Tian L, Nasir F, Ji L, Li X, Tian C. Comparative analysis of the rhizomicrobiome of the wild versus cultivated crop: insights from rice and soybean. Archives of Microbiology. 2019;201(7):879–888. doi: 10.1007/s00203-019-01638-8. [DOI] [PubMed] [Google Scholar]
- 40.Wissuwa M, Mazzola M, Picard C. Novel approaches in plant breeding for rhizosphere-related traits. Plant and Soil. 2009;321(1-2):409–430. [Google Scholar]
- 41.Zhang A, Shi H, Xu K, Sun Y, H, Lei J, M. Chemotaxis response of rhizosphere solani and sclerotinia schinseng to total ginsenosideds. Journal of Northwest A&F University. 2016;44(05):200-204+214. [Google Scholar]
- 42.Zhan Y, Wang E, Wang H, Chen X, Meng X, Li Q, Chen C. Allelopathic effects of ginsenoside on soil sickness, soil enzymes, soil disease index and plant growth of Ginseng. Allelopathy Journal. 2021;52:251–260. [Google Scholar]
- 43.Choi YE, Kim YS, Yi MJ, Park WG, Yi JS, Chun SR, Han SS, Lee SJ. Physiological and chemical characteristics of field-and mountain-cultivated ginseng roots. Journal of Plant Biology. 2007;50(2):198–205. [Google Scholar]
- 44.Ying YX, Ding WL, Li Y. Characterization of soil bacterial communities in rhizospheric and nonrhizospheric soil of panax ginseng. Biochem Genet. 2012;50(11-12):848–859. doi: 10.1007/s10528-012-9525-1. [DOI] [PubMed] [Google Scholar]
- 45.Szoboszlay M, Lambers J, Chappell J, Kupper JV, Moe LA, McNear DH. Comparison of root system architecture and rhizosphere microbial communities of Balsas teosinte and domesticated corn cultivars. Soil Biology and Biochemistry. 2015;80:34–44. [Google Scholar]
- 46.Voriskova J, Baldrian P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME Journal. 2013;7(3):477–486. doi: 10.1038/ismej.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei X, Wang X, Cao P, Gao Z, Chen AJ, Han J. Microbial Community Changes in the Rhizosphere Soil of Healthy and Rusty Panax ginseng and Discovery of Pivotal Fungal Genera Associated with Rusty Roots. BioMed Research Int. 2020;2020(1):1-13. [DOI] [PMC free article] [PubMed]
- 48.Glinushkin AP, Ovsyankina AV, Kornyukov DA: Diagnosis of fungi of the genus Fusarium and Alternaria, Bipolaris, causing diseases of sunflower, and immunological methods for the evaluation and selection of genotypes to the pathogens. IOP Conference Series: Earth and Environmental Science 2021, 663(1):012049.
- 49.Okello PN, Petrovic K, Singh AK, Kontz B, Mathew FM. Characterization of species of Fusarium causing root rot of Soybean (Glycine max L.) in South Dakota, USA. Canadian Journal of Plant Pathology. 2020;42(4):560–571. [Google Scholar]
- 50.Punja ZK, Wan A, Goswami RS, Verma N, Rahman M, Barasubiye T, Seifert KA, Lévesque CA. Diversity of Fusarium species associated with discolored ginseng roots in British Columbia. Canadian Journal of Plant Pathology. 2007;29(4):340–353. [Google Scholar]
- 51.Wei X, Wang X, Cao P, Gao Z, Chen AJ, Han J. Microbial Community Changes in the Rhizosphere Soil of Healthy and Rusty Panax ginseng and Discovery of Pivotal Fungal Genera Associated with Rusty Roots. BioMed Res Int. 2020;2020:8018525. doi: 10.1155/2020/8018525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang W, Sun D, Fu J, Zhao H, Wang R, An Y. Effects of continuous sugar beet cropping on rhizospheric microbial communities. Genes. 2020;11(1):13. [DOI] [PMC free article] [PubMed]
- 53.Center. NESSD: Soil SubCenter, National Science & Technology Infrastructure of China. (http://soilgeodatacn) 2020.
- 54.Ullah A, Akbar A, Luo Q, Khan AH, Manghwar H, Shaban M, Yang X. Microbiome Diversity in Cotton Rhizosphere Under Normal and Drought Conditions. Microbial Ecol. 2019;77(2):429–439. doi: 10.1007/s00248-018-1260-7. [DOI] [PubMed] [Google Scholar]
- 55.Haichar FEZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008;2(12):1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- 56.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 58.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dixon P. VEGAN, a package of R functions for community ecology. J Vegetation Sci. 2003;14(6):927–930. [Google Scholar]
- 61.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pẽa AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing dataset analyzed during the current study is available in the NCBI Sequence Read Archive (PRJNA701796 and PRJNA701800).