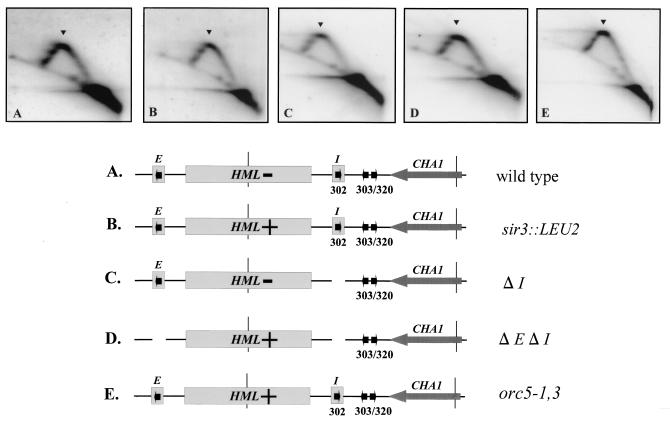
FIG. 7.
Detection of replication fork pause sites at the HML ARS cluster is independent of mutations that abrogate transcriptional silencing at HML, as determined by 2D gel electrophoresis of replication intermediates from the indicated strains probed for the 3.9-kb EcoRI/FspI fragment diagrammed in the maps. The maps in all panels show positions of the HML locus, the E and I silencer elements, and the ARS elements and the transcriptional status of HML (−, silent; +, active). A HindIII/BamHI fragment from was used as a probe in the hybridization. Arrowheads denote a replication pause signal. (A) DMY1, the parental strain; (B) DMY2, a strain with a mutation disrupting the SIR3 gene (shown as sir3::LEU2); (C) DMY94, a strain with the I silencer deleted (ΔI); (D) DMY95, a strain with E and I silencers deleted (ΔE ΔI); (E) JRY4556, a strain with a mutation in ORC5 (orc5-1,3) which is defective in transcriptional silencing but competent in DNA replication.