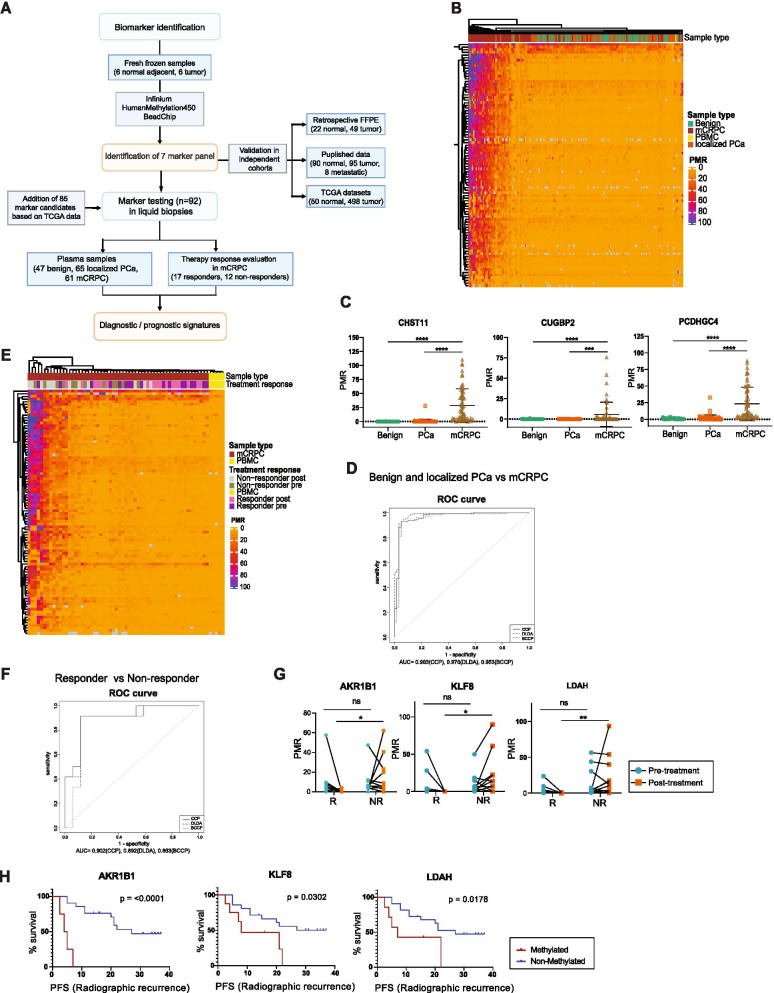
Fig. 1.
Identification and liquid biopsy testing of DNA methylation-based markers. A Experimental Workflow. Flow chart indicates biomarker identification, validation and analysis in different patient cohorts and publicly available datasets. B Unsupervised clustering of PMR-values resulting from MSRE-qPCR analysis of ccfDNA isolated from plasma of indicated patient groups for 92 marker candidates (n=47 benign, n=65 localized PCa, n=61 mCRPC, n=1 PBMC). C DNA methylation levels of ccfDNA isolated from plasma in patients with localized PCa or mCRPC versus controls for three signature genes. Differences between the three groups were assessed using one-way ANOVA (CHST11, PCDHGC4 n=47 benign, n=65 localized PCa, n=61 mCRPC, CUGBP2 n=46 benign, n=62 localized PCa, n=54 mCRPC; **** p < 0.0001). D ROC-curve analysis based on the three gene signature as in (C) for classification of mCRPC samples compared to benign and localized PCa patients combined, calculated with different prediction algorithms, ((Bayesian) Compound Covariate Predictor (BCCP, CCP) and Diagonal Linear Discriminant Analysis (DLDA)) using recursive feature elimination (n=112 benign + localized PCa, n=61 mCRPC). E Unsupervised clustering of PMR-values resulting from MSRE-qPCR of ccfDNA isolated from plasma of mCRPC patients before and after treatment, or healthy PBMC controls for 92 marker candidates (n=17 responders, n=12 non-responders, n=5 PBMC). F ROC-curve analysis based on a 3-gene signature (AKR1B1, KLF8, LDAH) for classification of responders versus non-responders. Calculations were performed using BCCP, CCP and DLDA with recursive feature elimination using BRB array tools software. G Methylation levels of the three signature genes for individual responders and non-responders pre- and post-treatment (n=17 responder, n=12 non-responder ** p < 0.01, * p < 0.05, ns p > 0.05; two-way ANOVA). H Kaplan-Meier-Analysis for radiographic progression-free survival (rPFS) of three signature genes using post-treatment samples (LDAH, KLF8 n=17 responder, n=12 non-responder; AKR1B1 n=15 responder, n=10 non-responder; p values shown on each plot calculated with Mantel-Cox-test, censored subjects indicated on plots by strokes)