Abstract
Over the last two decades, there have been three deadly human outbreaks of Coronaviruses (CoVs) caused by SARS-CoV, MERS-CoV, and SARS-CoV-2, which has caused the current COVID-19 global pandemic. All three deadly CoVs originated from bats and transmitted to humans via various intermediate animal reservoirs. It remains highly possible that other global COVID pandemics will emerge in the coming years, caused by yet another spillover of a bat-derived SARS-like Coronavirus (SL-CoV) into humans. Determining the antigen and the human B cells, CD4+ and CD8+ T cells epitope landscapes that are conserved among human and animal Coronaviruses should inform in the development of future pan-Coronavirus vaccines. In the present study, using several immuno-informatics and sequence alignment approaches, we identified several human B-cell, CD4+ and CD8+ T cell epitopes that are highly conserved in: (i) greater than 81,000 SARS-CoV-2 strains identified in 190 countries on six continents; (ii) six circulating CoVs that caused previous human outbreaks of the “Common Cold”; (iii) nine SL-CoVs isolated from bats; (iv) nine SL-CoV isolated from pangolins; (v) three SL-CoVs isolated from civet cats; and (vi) four MERS strains isolated from camels. Furthermore, we identified epitopes: (i) recalled B cell, CD4+ and CD8+ T cells from both COVID-19 patients and healthy individuals who were never exposed to SARS-CoV-2; and (ii) induced strong B cell and T cell responses in “humanized” Human Leukocyte Antigen (HLA)-DR1/HLA-A*02:01 double transgenic mice. The findings pave the way to develop a pre-emptive multi-epitope pan-Coronavirus vaccine to protect against past, current, and future outbreaks.
Keywords: SARS-CoV-2, SL-CoVs, COVID-19, Pan-Coronavirus, Vaccine, Epitopes, Antibodies, CD4+ T cells, CD8+ T cells, Immunity, Immunopathology
INTRODUCTION
As deforestation continues to expand and humans progressively conquer wildlife habitats around the globe, the wildlife “fights back” by spilling over many zoonotic viruses into human populations (1, 2). Among these, is the large family Coronaviruses. Since the first human Coronavirus was identified in 1965, many additional Coronavirus strains have continued to emerge (3–5). These caused several major human Coronavirus outbreaks within the last two decades (i.e., from 2002 to 2019): SARS-CoV (6); CoV-NL63 (7); CoV-HKU1 (8); CoV-229E (8); CoV-OC43 (9); MERS-CoV (10); and the highly contagious and deadly SARS-CoV-2 (11, 12). The many deadly Coronavirus outbreaks in the past twenty years should have been the impetus for urgently developing a pre-emptive pan-Coronavirus vaccine.
The first two deadly Coronaviruses, the MERS-CoV and the SARS-CoV, originated from bats, as natural hosts and reservoirs, and were transmitted to humans from intermediate animals namely camels and civet-cats, respectively (10, 13–16). The third deadly SARS-CoV-2 appears to be 96% identical to a bat SARS-like Coronavirus (SL-CoV) strain, termed Bat-CoV-RaTG13, and transmitted to humans from a yet-to-be determined intermediate animal (17, 18). While human-to-human spread of the “common cold” Coronaviruses occurs frequently, only rarely do animal-to-human Coronavirus transmissions occur (19). However, the highly contagious SARS-CoV-2 successfully produces both animal-to-human spread and human-to-human transmission (20, 21). The first known human-to-human transmission of SARS-CoV-2, which causes Coronavirus disease-2019 (COVID-19), was reported in late January 2020, prompting the WHO and US authorities to declare a global public health emergency (22).
All human Coronaviruses are associated with respiratory illnesses, ranging from mild common colds to more severe lower respiratory tract symptoms (23). Within 2–14 days after SARS-CoV-2 exposure, newly infected individuals may develop fever, fatigue, myalgia and respiratory symptoms including cough and shortness of breath (24). While 40–45% of newly infected individuals remained asymptomatic, 55–60% individuals are symptomatic ranging from mild/severe to critically ill patients, especially the elderly and those with comorbidities (24) (25). They develop severe pulmonary inflammatory disease and may need a rapid medical intervention to prevent acute respiratory distress syndrome and death (26, 27). The SARS-CoV-2 infection induces antiviral CD4+ T cells, helping the production of neutralizing/blocking antibodies and the formation of effector IFN-γ-producing CD4+ T cells and cytotoxic CD8+ T cells, all arms of immunity critical in reducing viral load in the majority of asymptomatic and convalescence patients (28–34). While SARS-CoV-2-specific IgG/IgM antibodies and CD4+ and CD8+ T cells are critical to reducing viral infection in a majority of asymptomatic and convalescence patients (35), an excessive proinflammatory cytokine storm appears to lead to acute respiratory distress syndrome and death in many symptomatic individuals (36–42). Thus, it is crucial to determine the B cell and T cell-epitope-specificities and the repertoire, the phenotype and function of B cells and CD4+ and CD8+ T cells that are associated with natural resistance seen in asymptomatic patients (43–45). The information herein should guide in the development of pan-Coronavirus vaccines.
In the present study, we identified several human B, CD4+ and CD8+ T cell epitopes that are highly conserved among six strains of Coronaviruses previously reported to infect humans and over 81,000 strains of SARS -CoV-2 that currently circulate in 190 countries on six continents. Moreover, as immune targets for pre-emptive pan-coronavirus vaccines, we identified the epitopes that are common among the above human Coronaviruses and twenty-five animal strains isolated from bats, pangolins, civet cats, and camels. We demonstrated the antigenicity of these epitopes in both SARS-CoV-2 patients and unexposed healthy individuals; and their immunogenicity in “humanized” Human Leukocyte Antigen (HLA)-DR1/HLA-A*02:01 double transgenic mice. Our findings pave the way for incorporating these highly conserved B-cell and T-cell epitopes in future pre-emptive multi-epitope pan-Coronavirus vaccines that would be expected to, not only protect against COVID-19, but also against subsequent global outbreaks.
MATERIALS & METHODS
Human study population:
Sixty-three COVID-19 patients and ten unexposed healthy individuals, who had never been exposed to SARS-CoV-2 or COVID-19 patients, were enrolled in this study (Table 1). Seventy-eight percent were non-White (African, Asian, Hispanic and others) and 22% were white. Forty-four percent were females, and 56% were males with an age range of 26–95 (median 62)
Table 1.
Clinical and demographic features of symptomatic and asymptomatic COVID-19 patients and unexposed healthy individuals enrolled in the study
| Patients Characteristics | Severe Symptoms (n = 9) | Moderate Symptoms (n = 11) | Mild Symptoms (n = 32) | Asymptomatic (n = 11) | Healthy Individuals (n = 10) | |
|---|---|---|---|---|---|---|
|
| ||||||
| Demographic features | Age | 62 (26–95) | 56 (24–91) | 62 (24–87) | 54 (22–78) | 51 (25–67) |
| Gender (Male/Female) | 5/4 (56%/44%) | 9/2 (82%/18%) | 21/11 (66%/34%) | 2/9 (18%/82%) | 6/4 (60%/40%) | |
| Race (% White/non-White) | 2/7 (22%/78%) | 1/10 (9%/91%) | 4/28 (12%/88%) | 5/6 (45%/55%) | 5/5 (50%/50%) | |
| HLA phenotype | HLA-A*0201 (+ve) | 4/9 (44%) | 5/11 (45%) | 14/32 (44%) | 4/11 (36%) | 10/10 (100%) |
| HLA-DRB1 (+ve) | 9/9(100%) | 11/11 (100%) | 32/32 (100%) | 11/11 (100%) | 10/10 (100%) | |
| Clinical parameters | BMI | 25.2 (20.3–57.9) | 26.5 (20.9–33.5) | 30.3 (21.1–46.5) | 29.3 (17.6–60.8) | - |
| Temperature/Fever/Chills | 98.4 (97.9–99.9) | 100.3 (97.7–102.8) | 99.1 (97.8–102.8) | 98.7 (97.7–102.5) | - | |
| Cough | 4 (44%) | 6 (55%) | 16 (50%) | 1 (9%) | - | |
| Shortness of Breath/Dyspnea | 7 (78%) | 7 (63%) | 21 (66%) | 1 (9%) | - | |
| Fatigue/Myalgia | 0 (0%) | 4 (36%) | 15 (47%) | 1 (9%) | - | |
| Headache | 4 (44%) | 6 (54%) | 16 (50%) | 1 (9%) | - | |
| ICU Admission | 9 (100%) | 11 (100%) | 2 (6%) | 0 (0%) | - | |
| Ventilator Support | 6 (67%) | 1 (9%) | 1 (3%) | 0 (0%) | - | |
| WBC | 10.9 (7.4–14.8) | 8 (6–29.8) | 7.1 (3.9–18.9) | 30.6 (4.9–60.8) | - | |
| RBC | 4.07 (2.97–5.92) | 4.04 (2.68–4.59) | 4.4 (2.69–5.41) | 7.1 (4.17–16.2) | - | |
| Hemoglobin (g/L) | 11.1 (8.3–16.2) | 12.1 (8.4–13.8) | 13.1 (8.1–16.9) | 4.4 (4.01–12.9) | - | |
| Comorbidities | Diabetes | 2 (22%) | 7 (64%) | 18 (56%) | 5 (46%) | - |
| Hypertension | 7 (78%) | 7 (64%) | 22 (69%) | 4 (36%) | - | |
| Cardiovascular disease | 1 (11%) | 2 (18%) | 4 (13%) | 1 (9%) | - | |
| CAD | 0 (0%) | 1 (9%) | 2 (6%) | 0 (0%) | - | |
| ESRD | 1 (11%) | 3 (27%) | 4 (13%) | 0 (0%) | - | |
| Asthma/COPD | 1 (11%) | 1 (9%) | 1 (3%) | 2 (18%) | - | |
| Obesity | 3 (33%) | 1 (9%) | 16 (50%) | 5 (46%) | - | |
| Cancer | 1 (11%) | 0 (0%) | 6 (19%) | 1 (9%) | - | |
Patients were scored on a scale of 1 to 4 and then classified into 3 groups of Symptomatic patients (Severe symptoms (i.e. ICU admission +/− Intubation or death), Moderate symptoms (i.e. ICU admission), Mild symptoms (i.e. Inpatient only) and Asymptomatic patients (i.e. infected patients but with no symptoms). Unexposed healthy individuals with no history of COVID-19 or contact with COVID-19 patients. Median values shown along with range; BMI: Body mass index, CAD: Coronary artery disease, ESRD: End stage renal disease, COPD: Chronic obstructive pulmonary disease
Detailed clinical and demographic characteristics of the COVID-19 patients and the unexposed healthy individuals with respect to age, gender, HLA-A*02:01 and HLA-DRB1 distribution, COVID-19 disease severity, comorbidity and biochemical parameters are presented in Table 1. None of the symptomatic patients were on anti-viral or anti-inflammatory drug treatments at the time of blood sample collections. The COVID-19 patients (n = 63) were divided into 4 groups depending on the severity of the symptoms: Group 1 that comprised of SARS-CoV-2 infected patients that never developed any symptoms or any viral diseases (i.e., asymptomatic patients) (n = 11); Group 2 with mild symptoms (i.e., Inpatient only, n = 32); Group 3 with moderate symptoms (i.e., ICU admission, n = 11) and Group 4 with severe symptoms (i.e., ICU admission +/− Intubation or death, n = 9). As expected, compared to the asymptomatic group, all of the 3 symptomatic groups (i.e., mild, moderate and severe) had higher percentages of comorbidities, including diabetes (22% to 64%), hypertension (64% to 78%), cardiovascular disease (11% to 18%) and obesity (9% to 50%) (Table 1). The final Group 5 was comprised of unexposed healthy individuals (controls), with no history of COVID-19 or contact with COVID-19 patients (n = 10) collected prior to 2019. All subjects were enrolled at the University of California Irvine under Institutional Review Board-approved protocols (IRB # 2020–5779). A written informed consent was received from all participants prior to inclusion in this study.
Sequence comparison among SARS-CoV-2 and previous Coronavirus strains:
We retrieved 81,963 human SARS-CoV-2 genome sequences from GISAID database representing countries from North America, South America, Central America, Europe, Asia, Oceania, and Africa (Fig. 1). Furthermore, the full-length sequences of SARS-CoV strains (SARS-CoV-2-Wuhan-Hu-1 (MN908947.3), SARS-CoV-Urbani (AY278741.1), HKU1-Genotype B (AY884001), CoV-OC43 (KF923903), CoV-NL63 (NC_005831), CoV-229E (KY983587)) and MERS (NC_019843)) found in the human host were obtained from the NCBI GenBank. SARS-CoV-2 genome sequences from bat (RATG13 (MN996532.2), ZXC21 (MG772934.1), YN01 (EPI_ISL_412976), YN02(EPI_ISL_412977)), and pangolin (GX-P2V (MT072864.1), GX-P5E (MT040336.1), GX-P5L (MT040335.1), GX-P1E (MT040334.1), GX-P4L (MT040333.1), GX-P3B (MT072865.1), MP789 (MT121216.1), Guangdong-P2S (EPI_ISL_410544)) were obtained from NCBI (www.ncbi.nlm.nih.gov/nuccore) and GSAID (www.gisaid.org). More so, the SARS-CoV strains from bat (WIV16 (KT444582.1), WIV1 (KF367457.1), YNLF_31C (KP886808.1), Rs672 (FJ588686.1), recombinant strain (FJ211859.1), camel (KT368891.1, MN514967.1, KF917527.1, NC_028752.1), and civet (Civet007, A022, B039)) were also retrieved from the NCBI GenBank. The sequences were aligned using ClustalW algorithm in MEGA X.
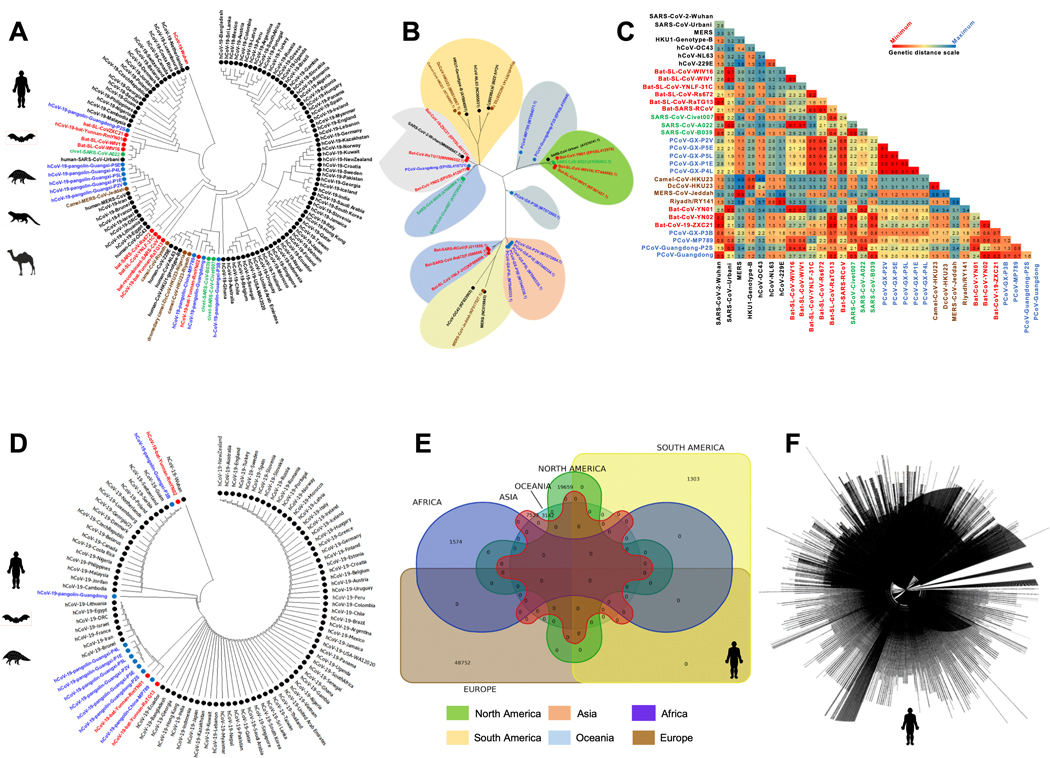
Figure 1. Evolutionary comparison of genome sequences among beta-Coronavirus strains isolated from humans and animals:
(A) Left panel: Phylogenetic analysis performed between SARS-CoV-2 strains (obtained from humans (Homo Sapiens (black)), along with the animal’s SARS-like Coronaviruses genome sequence (SL-CoVs) sequences obtained from bats (Rhinolophus affinis, Rhinolophus malayanus (red)), pangolins (Manis javanica (blue)), civet cats (Paguma larvata (green)), and camels (Camelus dromedaries (Brown)). The included SARS-CoV/MERS-CoV strains are from previous outbreaks (obtained from humans (Urbani, MERS-CoV, OC43, NL63, 229E, HKU1-genotype-B), bats (WIV16, WIV1, YNLF-31C, Rs672, recombinant strains), camel (Camelus dromedaries, (KT368891.1, MN514967.1, KF917527.1, NC_028752.1), and civet (Civet007, A022, B039)). The human SARS-CoV-2 genome sequences are represented from six continents. (B) Phylogenetic analysis performed among SARS-CoV-2 strains from human and other species with previous strains of SARS/MERS-CoV showed minimum genetic distance between the first SARS-CoV-2 isolate Wuhan-Hu-1 reported from the Wuhan Seafood market with bat strains hCoV-19-bat-Yunnan-RmYN02, bat-CoV-19-ZXC21, and hCoV-19-bat-Yunnan-RaTG13. This makes the bat strains nearest precursor to the human-SARS-CoV-2 strain. (C) Genetic distances based on Maximum Composite Likelihood model among the human, bat, pangolin, civet cat and camel genome sequences. Results indicate least genetic distance among SARS-CoV-2 isolate Wuhan-Hu-1 and bat strains bat-CoV-19-ZXC21 (0.1), hCoV-19-bat-Yunnan-RaTG13 (0.1), and hCoV-19-bat-Yunnan-RmYN02 (0.2). (D) Evolutionary analysis performed among the human-SARS-CoV-2 genome sequences reported from six continents and SARS-CoV-2 genome sequences obtained from bats (Rhinolophus affinis, Rhinolophus malayanus), and pangolins (Manis javanica)) (E) Venn diagram showing the number of SARS-CoV-2 genome sequences reported from Africa (n = 1574), Asia (n = 7533), North America (n = 19659), South America (n = 1303), Europe (n = 48752), and Oceania region (n = 3142) as on August 18, 2020. (F) Complete genome tree derived from 81,963 outbreak SARS-CoV-2 genome sequences submitted from Asian, African, North American, South American, European, and Oceanian regions.
Sequence conservation analysis of SARS-CoV-2:
The SARS-CoV-2-Wuhan-Hu-1 (MN908947.3) protein sequence was compared with SARS-CoV and MERS-CoV specific protein sequences obtained from human, bat, pangolin, civet and camel. The Sequence Variation Analysis was performed on the consensus aligned protein sequences from each virus strain. This Sequence Homology Analysis identified consensus protein sequences from the SARS-CoV and MERS-CoV and predicted the Epitope Sequence Analysis.
SARS-CoV-2 CD8 and CD4 T Cell Epitope Prediction:
Epitope prediction was carried out using the twelve proteins predicted for the reference SARS-CoV-2 isolate, Wuhan-Hu-1. The corresponding SARS-CoV-2 protein accession identification numbers obtained from NCBI (www.ncbi.nlm.nih.gov/protein) are: YP_009724389.1 (ORF1ab), YP_009725295.1 (ORF1a), YP_009724390.1 (surface glycoprotein), YP_009724391.1 (ORF3a), YP_009724392.1 (envelope protein), YP_009724393.1 (membrane glycoprotein), YP_009724394.1 (ORF6), YP_009724395.1 (ORF7a), YP_009725318.1 (ORF7b), YP_009724396.1 (ORF8), YP_009724397.2 (nucleocapsid phosphoprotein), YP_009725255.1 (ORF10). The tools used for CD8+ T cell-based epitope prediction were SYFPEITHI, MHC-I binding predictions, and Class I Immunogenicity. Of these, the latter two were hosted on the IEDB platform. For the prediction of CD4+ T cell epitopes, we used multiple databases and algorithms, namely SYFPEITHI, MHC-II Binding Predictions, Tepitool, and TEPITOPEpan. For CD8+ T cell epitope prediction, we selected the 5 most frequent HLA-A class I alleles (HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01, HLA-A*23:01) with large coverage of the world population, regardless of race and ethnicity (Supplemental Figs. S1A and S1C) (Middleton et al., 2003), using a phenotypic frequency cutoff ≥ 6%. Similarly, for CD4 T cell epitope prediction, selected HLA-DRB1*01:01, HLA-DRB1*11:01, HLA-DRB1*15:01, HLA-DRB1*03:01, HLA-DRB1*04:01 alleles with large population coverage (Supplemental Figs. S1B and S1D). Subsequently, using NetMHC we analyzed the SARS-CoV-2 protein sequence against all the aforementioned MHC-I and MHC-II alleles. Epitopes with 9mer length for MHC-I and 15mer length for MHC-II were predicted. Subsequently, the peptides were analyzed for binding stability to the respective HLA allotype. Our stringent epitope selection criteria were based on picking the top 1% epitopes focused on prediction percentile scores.
SARS-CoV-2 B Cell Epitope Prediction:
Linear B cell epitope predictions were carried out on the surface glycoprotein (S), the primary target of B cell immune responses for SARS-CoV. We used the BepiPred 2.0 algorithm embedded in the B cell prediction analysis tool hosted on IEDB platform. For each protein, the epitope probability score for each amino acid and the probability of exposure was retrieved. Potential B cell epitopes were predicted using a cutoff of 0.55 (corresponding to a specificity greater than 0.81 and sensitivity below 0.3) and considering sequences having more than 5 amino acid residues. This screening process resulted in 28 B-cell peptides (Supplemental Table S3). From this pool, we selected 10 B-cell epitopes with 19 to 62 amino acid lengths. Three B-cell epitopes were observed to possess receptor binding domain (RBD) region specific amino acids. Structure-based antibody prediction was performed by using Discotope 2.0, and a positivity cutoff greater than −2.5 was applied (corresponding to specificity greater than or equal to 0.80 and sensitivity below 0.39), using the SARS-CoV-2 spike glycoprotein structure (PDB ID: 6M1D).
Protein-peptide molecular docking:
Computational peptide docking of B cell peptides into the ACE2 Complex (binding protein) was performed using the GalaxyPepDock under GalaxyWEB. To retrieve the ACE2 structure, we used the X-ray crystallographic structure ACE2-B0AT1 complex-6M1D available on the Protein Data Bank. The 6M1D with a structural weight of 334.09 kDa, possesses 2 unique protein chains, 2,706 residues, and 21,776 atoms. In this study, flexible target docking based on an energy-optimization algorithm was carried out on the ligand-binding domain containing ACE2 within the 4GBX structure. Similarity scores were calculated for protein-peptide interaction pairs for each residue. The prediction accuracy is estimated from a linear model as the relationship between the fraction of correctly predicted binding site residues and the template-target similarity measured by the protein structure similarity score and interaction similarity score (SInter) obtained by linear regression. SInter shows the similarity of amino acids of the B-cell peptides aligned to the contacting residues in the amino acids of the ACE2 template structure. Higher SInter score represents a more significant binding affinity among the ACE2 molecule and B-cell peptides. Subsequently, molecular docking models were built based on distance restraints for protein-peptide pairs using GalaxyPepDock. Based on the optimized energy scores, docking models were ranked.
While performing the protein-peptide docking analysis for CD8+ T cell epitope peptides, we used the X-ray Crystal structure of HLA-A*02:01 in complex-4UQ3 available on the Protein Data Bank and for CD4 peptides X-ray crystallographic structure HLA-DM-HLA-DRB1 Complex-4GBX.
Epitope conservancy analysis:
The Epitope Conservancy Analysis tool was used to compute the degree of the conservancy of CD8+ T cell, CD4+ T cell, and B-cell epitopes within a given protein sequence of SARS-CoV-2 set at 100% identity level. The fraction of protein sequences that contain the regions similar to epitopes were evaluated on the degree of similarity or correspondence among two sequences. The CD8+ T cell, and CD4+ T cell epitopes were screened against all the twelve structural and non-structural proteins of SARS-CoV-2 namely YP_009724389.1 (ORF1ab), YP_009725295.1 (ORF1a), YP_009724390.1 (surface glycoprotein), YP_009724391.1 (ORF3a), YP_009724392.1 (envelope protein), YP_009724393.1 (membrane glycoprotein), YP_009724394.1 (ORF6), YP_009724395.1 (ORF7a), YP_009725318.1 (ORF7b), YP_009724396.1 (ORF8), YP_009724397.2 (nucleocapsid phosphoprotein), YP_009725255.1 (ORF10). B-cell epitopes were screened for their conservancy against surface glycoprotein (YP_009724390.1) of SARS-CoV-2. Epitope linear sequence conservancy approach was used for linear epitope sequences with a sequence identity threshold set at ≥ 50%. This analysis resulted in (i) the calculated degree of conservancy (percent of protein sequence matches a specified identity level) and (ii) the matching minimum/maximum identity levels within the protein sequence set. The CD8+ and CD4+ T cell epitopes that showed ≥ 50% conservancy in at-least two human SARS-CoV strains, and two SARS-CoV strains (from bat/civet/pangolin/camel) were selected as candidate epitopes. N and O glycosylation sites were screened using NetNGlyc 1.0 and NetOGlyc 4.0 prediction servers, respectively (46).
Population-Coverage-Based T Cell Epitope Selection:
For a robust epitope screening, we evaluated the conservancy of CD8+ T cell, CD4+ T cell, and B cell epitopes within Human-SARS-CoV-2 genome sequences representing North America, South America, Africa, Europe, Asia, and Australia. As of August 27th, 2020, the NextStrain database recorded 81,963 human-SARS-CoV-2 genome sequences and the number of genome sequences continues growing daily. In the present analysis, 81,963 human-SARS-CoV-2 genome sequences were extrapolated from the GISAID and NCBI GenBank databases. We therefore considered all the 81,963 SARS-CoV-2 genome sequences representing six continents for subsequent conservancy analysis. We set a threshold for a candidate CD8+ T cell, CD4+ T cell, and B-cell epitope if the epitope showed 100% sequence conservancy in ≥ 95 human-SARS-CoV-2 genome sequences. Furthermore, population coverage calculation (PPC) was carried out using the Population Coverage software hosted on IEDB platform (47). PPC was performed to evaluate the distribution of screened CD8+ and CD4+ T cell epitopes in world population at large in combination with HLA-I (HLA-A*01:01,HLA-A*02:01,HLA-A*03:01,HLA-A*11:01,HLA-A*23:01), and HLA-II (HLA-DRB1*01:01, HLA-DRB1*11:01, HLA-DRB1*15:01, HLA-DRB1*03:01, HLA-DRB1*04:01) alleles.
Peptide synthesis:
Potential peptide epitopes (9-mer long for CD8+ T cell epitopes and 15-mer long for CD4+ T cell epitopes) identified from twelve human-SARS-CoV-2 proteins namely ORF1ab, ORF1a, surface glycoprotein, ORF3a, envelope protein, membrane glycoprotein, ORF6, ORF7a, ORF7b, ORF8, nucleocapsid phosphoprotein, and ORF10 were synthesized using solid-phase peptide synthesis and standard 9-fluorenylmethoxycarbonyl technology (21st Century Biochemicals, Inc, Marlborough, MA). The purity of peptides was over 90%, as determined by reversed-phase high-performance liquid chromatography (Vydac C18) and mass spectroscopy (VOYAGER MALDI-TOF System). Stock solutions were made at 1 mg/mL in 10% DMSO in PBS. Similar method of synthesis was used for B cell peptide epitopes from the spike protein of SARS-CoV-2.
Cell Lines:
T2 (174 × CEM.T2) mutant hybrid cell line derived from the T-lymphoblast cell line CEM was obtained from the ATCC (www.atcc.org). The T2 cell line was maintained in IMDM (ATCC, Manassas, VA) supplemented with 10% heat-inactivated fetal calf serum (FCS) and 100 U of penicillin/mL, 100 U of streptomycin/mL (Sigma-Aldrich, St. Louis, MO).T2 cells lack the functional transporter associated with antigen processing (TAP) heterodimer and failed to express normal amounts of HLA-A*02:01 on the cell surface. HLA-A*02:01 surface expression is stabilized following the binding of exogenous peptides to these MHC class I molecules.
Stabilization of HLA-A*02:01 on class-I-HLA-transfected B x T hybrid cell lines:
To determine whether synthetic peptides could stabilize HLA-A*02:01 molecule expression on the T2 cell surface, peptide-inducing HLA-A*02:01 up-regulation on T2 cells was examined according to a previously described protocol (48, 49). T2 cells (3 × 105/well) were incubated with different concentrations (30, 10 and 3 μM) of 91 individual CD8+ T cell specific peptides in 48-well plates for 18 hours at 26°C. Cells were then incubated at 37°C for 3 hours in the presence of 0.7 μL/mL BD GolgiStop™ to block cell surface expression of newly synthesized HLA-A*02:01 molecules, and human β−2 microglobulin (1μg/mL). The cells were subsequently washed with FACS buffer (1% BSA and 0.1% sodium azide in phosphate-buffered saline) and stained with anti-HLA-A2 specific monoclonal antibody (clone BB7.2) (BD-Pharmingen, San Diego, CA) at 4°C for 30 minutes. After incubation, the cells were washed with FACS buffer, fixed with 2% paraformaldehyde in phosphate-buffered saline, and analyzed by flow cytometry using a Fortessa (Becton Dickinson) flow cytometer equipped with a BD High Throughput Sampler for rapid analysis of samples prepared in plate format. The acquired data were analyzed with FlowJo software (BD Biosciences, San Jose, CA) and expression was measured by mean fluorescence intensity (MFI). Percent MFI increase was calculated as follows: Percent MFI increase = (MFI with the given peptide - MFI without peptide) / (MFI without peptide) x 100. Each experiment was performed 3 times, and means ± SD values were calculated.
HLA-A*02:01 and HLA-DR1 double transgenic mice:
A colony of human leukocyte antigens (HLA) class I and class II double transgenic (Tg) mice was maintained at the University of California Irvine (50) vivarium and treated in accordance with the AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) according to Institutional Animal Care and Use Committee-approved animal protocols (IACUC # 2020–19-111), and NIH (National Institutes of Health) guidelines. The HLA Tg mice retain their endogenous mouse major histocompatibility complex (MHC) locus and express human HLA-A*02:01 and HLA-DRB*01 under the control of its normal promoter (51, 52). Prior to this study, the expression of HLA-A*02:01 and DR1 molecules on the PBMCs of each HLA-Tg mouse was confirmed by fluorescence-activated cell sorting (FACS).
Immunization of mice:
Groups of age-matched HLA transgenic mice/B6 mice (n = 3) were immunized subcutaneously, on days 0 and 14, with a mixture of four SARS-CoV-2-derived human CD4+ T/ CD8+T /B cell peptide epitopes delivered in alum and CpG1826 adjuvants. As a negative control, mice received adjuvants alone (mock-immunized).
Splenocytes isolation:
Spleens were harvested from mice in two weeks post second immunization. Spleens were placed in 10 ml of cold PBS with 10% fetal bovine serum (FBS) and 2X antibiotic–antimycotic (Life Technologies, Carlsbad, CA). Spleens were minced finely and sequentially passed through a 100 μm screen and a 70 μm screen (BD Biosciences, San Jose, CA). Cells were then pelleted via centrifugation at 400 × g for 10 minutes at 4°C. Red blood cells were lysed using a lysis buffer (ammonium chloride) and washed again. Isolated splenocytes were diluted to 1 × 106 viable cells per ml in RPMI media with 10% (v/v) FBS and 2 × antibiotic–antimycotic. Viability was determined by trypan blue staining.
Flow cytometry analysis:
PBMCs/Splenocytes were analyzed by flow cytometry. The following antibodies were used: CD8, CD4, CD62L, CD107a/b, CD44, CD69, TNF-α and IFN-γ). For surface staining, mAbs against various cell markers were added to a total of 1X106 cells in phosphate-buffered saline containing 1% FBS and 0.1% sodium azide (fluorescence-activated cell sorter [FACS] buffer) and left for 45 minutes at 4°C. At the end of the incubation period, the cells were washed twice with FACS buffer. A total of 100,000 events were acquired by LSRII (Becton Dickinson, Mountain View, CA) followed by analysis using FlowJo software (TreeStar, Ashland, OR).
ELISpot assay:
All reagents used were filtered through a 0.22 μm filter. Wells of 96-well Multiscreen HTS Plates (Millipore, Billerica, MA) were pre-wet with 30% ethanol and then coated with 100 μl primary anti-IFN-γ antibody solution (10 μg/ml of 1-D1K coating antibody from Mabtech in PBS, pH 7.4, V-E4) OVN at 4°C. After washing, nonspecific binding was blocked with 200 μl of RPMI media with 10% (v/v) FBS for 2 hours at room temperature. Following the blockade, 0.5 × 106 cells from patients PBMCs (or from mouse splenocytes) in 100 μl of RPMI were mixed with 10 μg individual peptides (with DMSO for no stimulation or with individual peptide at a final concentration of 10 μg/ml). After incubation in humidified 5% CO2 at 37°C for 72 hours (samples from COVID-19 patients) or 5 days (for healthy donor samples, to recall their T-cell memory), cells were removed by washing (using PBS and PBS-Tween 0.02% solution) and 100 μl of biotinylated secondary anti-IFN-γ antibody (clone 7-B6–1, Mabtech) in blocking buffer (PBS 0.5% FBS) was added to each well. Following a 2-hour incubation and washing, HRP-conjugated streptavidin was diluted 1:1000, and wells were incubated with 100 μl for 1 hour at room temperature. Following washing, wells were incubated for 1 hour at room temperature with 100 μl of TMB detection reagent and spots counted with an automated EliSpot Reader System (ImmunoSpot reader, Cellular Technology, Shaker Heights, OH).
ELISA based assay to access the efficacy of receptor-binding domain region towards inducing specific antibodies against B-cell epitopes in HLA-A2 treated mice:
The efficacy of our B-cell peptide-epitopes towards inducing specific antibodies was measured in the HLADR1/A*02:01 immunized mice by ELISA. ELISA plates (Cat. M5785, Sigma Aldrich) were first coated overnight at 4°C with 10μg/ml of each B cell peptide epitope. Subsequently, plates were washed five times with PBS-Tween 0.01% before starting the blocking by adding PBS 1% BSA for 3 hours at room temperature, followed by a second wash. Sera of C57BL/6 mice immunized either with pool B cell peptides alum/CpG or adjuvant alone (control) were added into the wells at various dilutions (1/5, 1/25, 1/125, and 1/625 or PBS only, in triplicates). Plates were incubated at 4°C overnight with the sera, then washed with PBS-Tween 0.01% before to add anti-mouse IgG antibody (Mabtech – 1/500 dilution). After the last washing, Streptavidin-HRP (Mabtech – 1/1000 dilution) was added for 30 minutes at room temperature. Finally, we added 100μl of filtered TMB substrate for 15 minutes and blocked the reaction with H2S04 before the read-out (OD measurement was done at 450nm on the Bio-Rad iMark microplate reader). The same procedure was followed to measure the titers of antibodies specific against our 15 screened B-cell epitopes in the sera of COVID-19 patients (n = 40) and healthy donors (n = 10), using anti-human IgG antibody as the secondary antibody (Mabtech – 1/500 dilution).
Constructing the Phylogenetic Tree:
Phylogenetic analyses were conducted in MEGA X. The evolutionary history was performed, and phylogenetic tree was constructed using the Maximum Likelihood method and Tamura-Nei model. The Maximum Likelihood method assumes that each locus evolves independently by pure genetic drift. The tree with the highest log likelihood was selected. Initial tree(s) for the heuristic search were obtained by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise genetic distances estimated using the Tamura-Nei model, and then selecting the topology with superior log likelihood value. This analysis involved available nucleotide sequences of SARS-CoV-2 from human (Homo Sapiens), bat (Rhinolophus affinis, Rhinolophus malayanus), and pangolin (Manis javanica). In addition, genome sequences from previous outbreaks of SARS-CoV in human, bat, civet, and camel were taken into consideration while performing the evolutionary analyses.
Data and Code Availability:
The human specific SARS-CoV-2 complete genome sequences were retrieved from the GISAID database, whereas the SARS-CoV-2 sequences for pangolin (Manis javanica), and bat (Rhinolophus affinis, Rhinolophus malayanus) were retrieved from NCBI. Genome sequences of previous strains of SARS-CoV for human, bat, civet, and camel were retrieved from the NCBI GenBank.
Statistical analyses:
Data for each differentially expressed markers among blockade-treated, and mock-treated groups of HLA Tg mice were compared by analysis of variance (ANOVA) and Student’s t-test using GraphPad Prism version 6 (La Jolla, CA). Statistical differences observed in the measured CD8-, CD4- T cells and antibody responses between healthy donors and COVID-19 patients were calculated using ANOVA and multiple t-test comparison procedures in GraphPad Prism. Data are expressed as the mean ± SD. Results were considered statistically significant at P ≤ 0.05.
RESULTS
1. Evolutionary convergence of human SARS-CoV-2 into bat and pangolin-derived SARS-like Coronaviruses:
Understanding the animal origins of SARS-CoV-2 is critical for the development of a pre-emptive pan-Coronavirus vaccine to protect from future human outbreaks and deter future zoonosis.
We first screened for the evolutionary relationship among human SARS-CoV-2 and SARS-CoV/MERS-CoV strains from previous outbreaks (i.e., Urbani, MERS-CoV, OC43, NL63, 229E, HKU1-genotype-B) along with 25 SARS-like Coronaviruses genome sequence (SL-CoVs) obtained from different animal species: Bats (Rhinolophus affinis and Rhinolophus malayanus), civet cats (Paguma larvata) and pangolins (Manis javanica), and MERS-CoVs from camels (Camelus dromedarius and Camelus bactrianus) (Fig. 1). These sequence alignments revealed similarity of the original human-SARS-CoV-2 strain found in Wuhan, China to four bat SL-CoV strains: hCoV-19-bat-Yunnan-RmYN02, bat-CoV-19-ZXC21, and hCoV-19-bat-Yunnan-RaTG13 obtained from the Yunnan and Zhejiang provinces of China (Fig. 1A). With further genetic distance analysis, we discovered the least evolutionary divergence between SARS-CoV-2 isolate Wuhan-Hu-1 and the above mentioned three SL-CoVs isolates from bats, namely: (1) Bat-CoV-RaTG13 (0.1), (2) bat-CoV-19-ZXC21 (0.1) and (3) Bat-CoV-YN02 (0.2) (Fig. 1B and 1C). Moreover, the phylogenetic analysis performed among the whole genome sequences of a total of 81,963 SARS-CoV-2 strains for which sequences have been reported in circulation in 190 countries suggest an evolutionary convergence of bat and pangolin SL-CoVs into the human SARS-CoV-2 strains (Figs. 1D and 1E). Furthermore, through a complete genome tree derived from the 81,963 SARS-CoV-2 genome sequences submitted from Asian, African, North American, South American, European, and Oceanian regions, we confirmed that the least evolutionary divergence for SARS-CoV-2 strains is in SL-CoVs isolated from bats and pangolins (Figs. 1D, 1E and 1F).
Altogether, the phylogenetic analysis and genetic distance suggest that the highly contagious and deadly human-SARS-CoV-2 strain originated from bats, most likely from either the Bat-CoV-19-ZXC21 (MG772934.1) or Bat-CoV-RaTG13 (MN996532.2) strains, that spilled over into humans after further mutations and/or recombination. These mutations and/or recombination(s) possibly contributed to the rapid global expansion of the highly contagious and deadly SARS-CoV-2 (53, 54).
2. Genome-wide identification of SARS-CoV-2 CD8+ T cell epitopes that are highly conserved between human and bat/pangolin Coronaviruses:
We first predicted potential CD8+ T cell epitopes from the entire genome sequence of the first SARS-CoV-2-Wuhan-Hu-1 strain (NCBI GenBank accession number MN908947.3) (55–61). For this, we used multiple databases and algorithms including the SYFPEITHI, MHC-I processing predictions, MHC-I binding predictions, MHC-I immunogenicity and Immune Epitope Database (IEDB) (58, 62). We focused on epitopes restricted to the five most frequent human leukocyte antigen (HLA) class I alleles with large coverage in worldwide human populations, regardless of race and ethnicity (i.e., HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01, HLA-A*23:01) (63–65) (Supplemental Figs. S1A and S1C).
Using the aforementioned criteria, we originally identified a total of 9,660 potential CD8+ T cell epitopes derived from 12 structural proteins (surface glycoprotein, membrane glycoprotein, nucleocapsid phosphoprotein) and open-reading-frames (ORFs) of SARS-CoV-2-Wuhan-Hu-1 strain (MN908947.3) (Supplemental Table S1). Subsequently, this large pool of epitopes was narrowed down to 91 epitopes, that are highly conserved among: (i) over 81,000 SARS-CoV-2 strains (that currently circulate in 190 countries on 6 continents); (ii) the 4 major “common cold” Coronaviruses that caused previous outbreaks (i.e., hCoV-OC43 (KF923903), hCoV-229E (KY983587), hCoV-HKU1 genotype B (AY884001), and hCoV-NL63 (NC_005831)); and (iii) the SL-CoVs that are isolated from bats, civet cats, pangolins and camels (Fig. 2A). While the highest degree of similarity (expressed as % of resemblance) was identified among 81,963 SARS-CoV-2 strains, 6 strains of previous human SARS-CoVs and 18 animal SL-CoVs strains isolated from bats and pangolins, only a small percentage of similarity was found between the SARS-CoV-2 and MERS-CoV strains (Supplemental Fig. 2). However, a significantly lower degree of similarity was recorded amongst the SARS-CoV-2 and the SL-CoVs strains isolated from civet cats’ and camels’ CoVs (Supplemental Fig. 2).
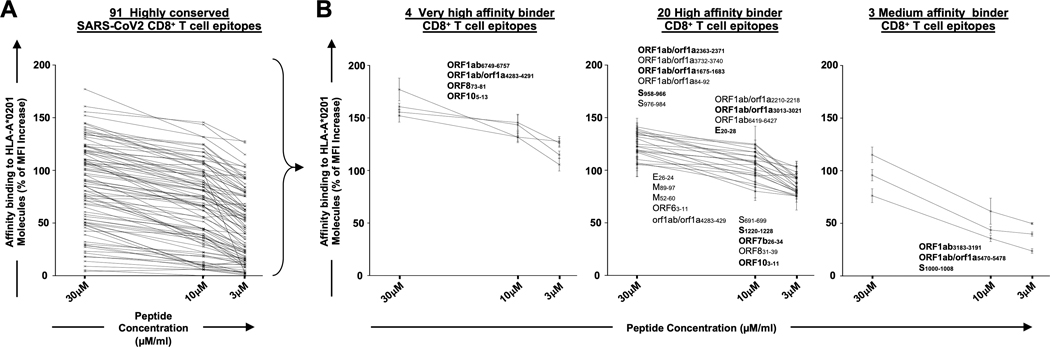
Figure 2: Identification of highly conserved potential SARS-CoV-2-derived human CD8+ T cell epitopes that bind with high affinity to HLA-A*02:01 molecules:
(A) Ninety-one, genome-wide In-silico predicted, and highly conserved SARS-CoV-2-derived CD8+ T cell epitope peptides were synthetized and were tested for their binding affinity in vitro to HLA-A*02:01 molecules expressed on the surface of T2 cells. (B) Out of the 91 CD8+ T cell epitopes, 4 epitopes were selected as high binders s to HLA-A*02:01 molecules, even at the lowest molarity of 3 uM. Further, 20 epitopes with high and 3 epitopes with moderate binding affinity found to stabilize the expression of HLA- A*02:01 molecules on the surface of the T2 cells. The levels of HLA-A*02:01 surface expression was determined by mean fluorescence intensity (MFI), measured by flow cytometry on T2 cells following an overnight incubation of T2 cells at 26°C with decreasing peptide epitopes molarity (30, 15 and 5μM) as shown in graphs. Percent MFI increase was calculated as follows: Percent MFI increase = (MFI with the given peptide - MFI without peptide) / (MFI without peptide) X 100.
We further identified 27 SARS-CoV-2 human CD8+ T cell epitopes, out of the 91 epitopes, that bound with high affinity with HLA-A*02:01 molecules, using in vitro peptide-HLA binding assay (Fig. 2A). Four epitopes were found to be very high affinity binders (Fig. 2B). The 27 epitopes with high binding affinity were later confirmed in silico using molecular docking models across 5 major HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01, HLA-A*23:01 haplotypes (Supplemental Fig. S3) (66). The highest binding affinity to HLA-A*02:01 molecules, with the highest interaction similarity (Sinter) scores (blue squares), were recorded for ORF1ab6749–6757, S2–10, S958–966, S1220–1228, E26–34, ORF883–91, ORF103–11 and ORF105–13 whereas minimum Sinter score was observed for ORF1ab3732–3740, S691–699 and M89–97. Other CD8+ T cell epitopes like ORF1ab1675–1683, ORF1ab2363–2371, ORF1ab3013–3021 and ORF7b26–34 were also found with intermediate Sinter scores (Supplemental Figs. S3A and S3B). While the identified highly conserved CD8+ T cell epitopes were distributed within 8 of the 12 structural and non-structural ORFs (i.e., ORF1ab, S, E, M, ORF6, ORF7b, ORF8, and ORF10), the highest numbers of epitopes were localized in the replicase polyprotein 1ab/1a (ORF1ab) (9 epitopes) followed by the spike glycoprotein (S) (5 epitopes) (Supplemental Fig. S2 and Fig. 8).
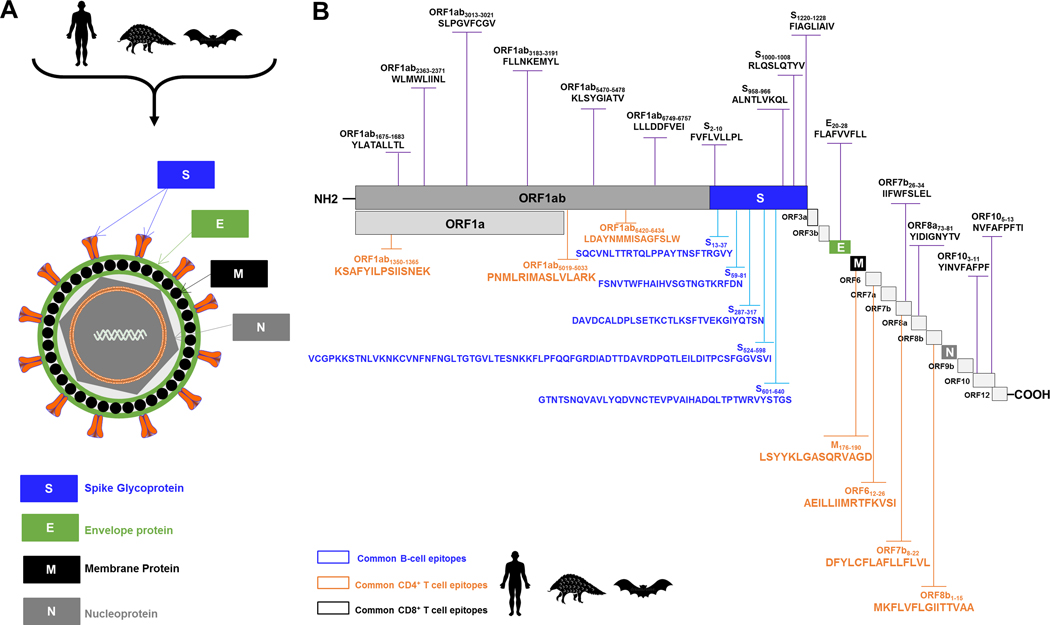
Figure 8: Illustrations of the SARS-CoV/SARS-CoV-2 genome-wide location of the highly conserved, antigenic and immunogenic CD4+ T cell, CD8+ T cell, and B-cell epitopes.
(A) Enveloped, spherical, about 120 nm in diameter, SARS-CoV/SARS-CoV-2 genome encodes four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N), highlighted in blue, green, gray and black, respectively. (B) The SARS-CoV/SARS-CoV-2 genome encodes two large non-structural genes ORF1a (green) and ORF1b (gray), encoding 16 non-structural proteins (NSP1– NSP16). The genome encodes at least six accessory proteins (shades of light grey) that are unique to SARS-CoV/SARS-CoV-2 in terms of number, genomic organization, sequence, and function. The common SARS-CoV, SARS-CoV-2 and SL-CoVs-derived human B (blue), CD4+ (green) and CD8+ (black) T cell epitopes are shown. Structural and non-structural open reading frames utilized in this study were from SARS-CoV-2-Wuhan-Hu-1 strain (NCBI accession number MN908947.3). The amino acid sequence of the SARS-CoV-2-Wuhan-Hu-1 structural and non-structural proteins was screened for human B, CD4+ and CD8+ T cell epitopes using different computational algorithms as previously described in Materials and Methods. Shown are genome-wide identified SARS-CoV-2 human B cell epitopes (in blue), CD4+ T cell epitopes (in green), CD8+ T cell epitopes (in black) that are highly conserved between human and animal Coronaviruses.
Altogether, our findings identified 27 highly conserved potential human CD8+ T cell epitopes from the sequence of SARS-CoV-2 that are highly conserved among 81,963 SARS-CoV-2 strains, the 4 major “common cold” Coronaviruses (i.e., hCoV-OC43, hCoV-229E, hCoV-HKU1 genotype B, and hCoV-NL63), newly found highly transmissible variants (Supplementary Fig. S9) and several SL-CoV strains that are isolated from bats and pangolins. These results suggest that both the structural and the non-structural proteins are immunodominant antigens that are targeted by human CD8+ T cells from both COVID-19 patients and “common cold” Coronaviruses infected healthy individuals.
3. In silico screening of potential promiscuous SARS-CoV-2 CD4+ T cell epitopes that are highly conserved between human and bat/pangolin Coronaviruses:
We subsequently identified a total of 9,594 potential HLA-DR-restricted CD4+ T cell epitopes from the whole genome sequence of SARS-CoV-2-Wuhan-Hu-1 strain (MN908947.3) using multiple databases and algorithms including the SYFPEITHI, MHC-II Binding Predictions, Tepitool and TEPITOPEpan (Supplemental Table S2). These potential promiscuous CD4+ T cell epitopes were screened in silico against the five most frequent HLA-DR alleles with large coverage in the human population, regardless of race or ethnicity: HLA-DRB1*01:01, HLA-DRB1*11:01, HLA-DRB1*15:01, HLA-DRB1*03:01, HLA-DRB1*04:01 (Supplemental Figs. S1B and S1D). The number of potential CD4+ T cell epitopes was later narrowed down to 16 epitopes based on: (i) the epitope sequences that are highly conserved among 81,963 SARS-CoV-2 strains, the 4 major “common cold” and 25 SL-CoV strains isolated from bats, civet cats, pangolins and camels (Supplemental Fig. S4); and (ii) their high binding affinity to HLA-DR molecules using in silico molecular docking models (Supplemental Fig. S5). The sequences of most of the 16 CD4+ T cell epitopes are 100% conserved and common among 81,963 SARS-CoV-2 strains currently circulating in 6 continents (Supplemental Fig. S4). A high degree of sequence similarities was also identified in the sequences of most 16 CD4+ T cell epitopes among the SARS-CoV-2 strains and the six strains of previous human SARS-CoVs (e.g., up to 100 % sequence identity for epitopes ORF1ab5019–5033, ORF1ab6088–6102, ORF1ab 6420–6434, E20–34, E26–40 and M176–190). Moreover, a high degree of sequence similarities was also identified among the SARS-CoV-2 and the SL-CoV strains isolated from bats and pangolins. In contrast, a lower sequence similarity, was identified among CD4+ T cell epitopes from SARS-CoV-2 strains and the SL-CoV strains isolated from civet cats followed by MERS-like CoV strains isolated from camels (Supplemental Fig. S4 and Fig. 8).
The 16 highly conserved CD4+ T cell epitopes are distributed within 9 out of the 12 structural and non-structural ORFs (i.e., ORF1ab, S, E, M, ORF6, ORF7a, ORF7b, ORF8 and N). The highest numbers of epitopes were localized in the replicase polyprotein ORF1ab/1a (5 epitopes) followed by ORF7a (3 epitopes) (Supplemental Fig. S4 and Fig. 8). Unlike the human CD8+ T cell epitopes, the human CD4+ T cell epitopes are found to be expressed in each of the structural S, E, M, and N proteins. Two epitopes are from the envelope protein (E), 1 epitope from the membrane protein (M), 1 epitope from the nucleoprotein (N) protein, and 1 epitope from the spike protein (S). The remaining CD4+ T cell epitopes are distributed among the ORF6, ORF7a, ORF7b and ORF8 proteins (Supplemental Fig. S4 and Fig. 8).
Altogether, these results identified 16 potential CD4+ T cell epitopes from the whole sequence of SARS-CoV-2 that cross-react and have high sequence similarity among 81,963 SARS-CoV-2 strains, the main 4 major “common cold” Coronaviruses and the SL-CoV strains isolated from bats and pangolins. Similar to CD8+ T cell epitopes, the replicase polyprotein ORF1ab appeared to be the most immunodominant antigen with a high number of conserved epitopes that may possibly be targeted by human CD4+ T cells.
4. Cross-reactive human and animal Coronavirus-derived epitopes, spanning the whole virus proteome, are targeted by memory CD4+ and CD8+ T cells from SARS-CoV-2 patients and unexposed healthy individuals:
Next, we assessed whether the potential SARS-CoV-2 CD4+ and CD8+ T cell epitopes that are highly conserved between human and animal Coronaviruses would recall memory CD8+ T cells from COVID-19 patients as well as from healthy individuals, who have never been exposed to SARS-CoV-2 or to COVID-19 patients (i.e., from healthy individuals blood samples that were collected from 2014 to 2018, Figs. 3A and 4A). Detailed clinical and demographic characteristics of the COVID-19 patients and the unexposed healthy individuals enrolled in the present study, with respect to age, gender, HLA-A*02:01 and HLA-DRB1 distribution, COVID-19 disease severity, comorbidity and biochemical parameters are described in Table 1 and in Materials and Methods.
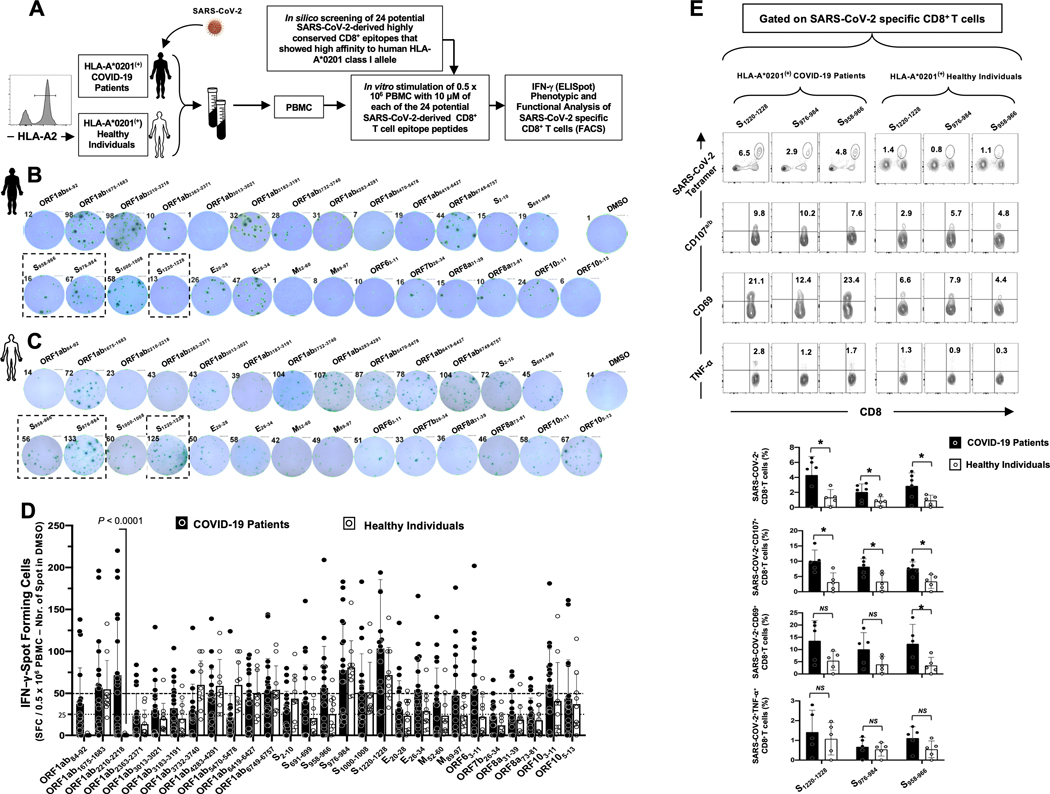
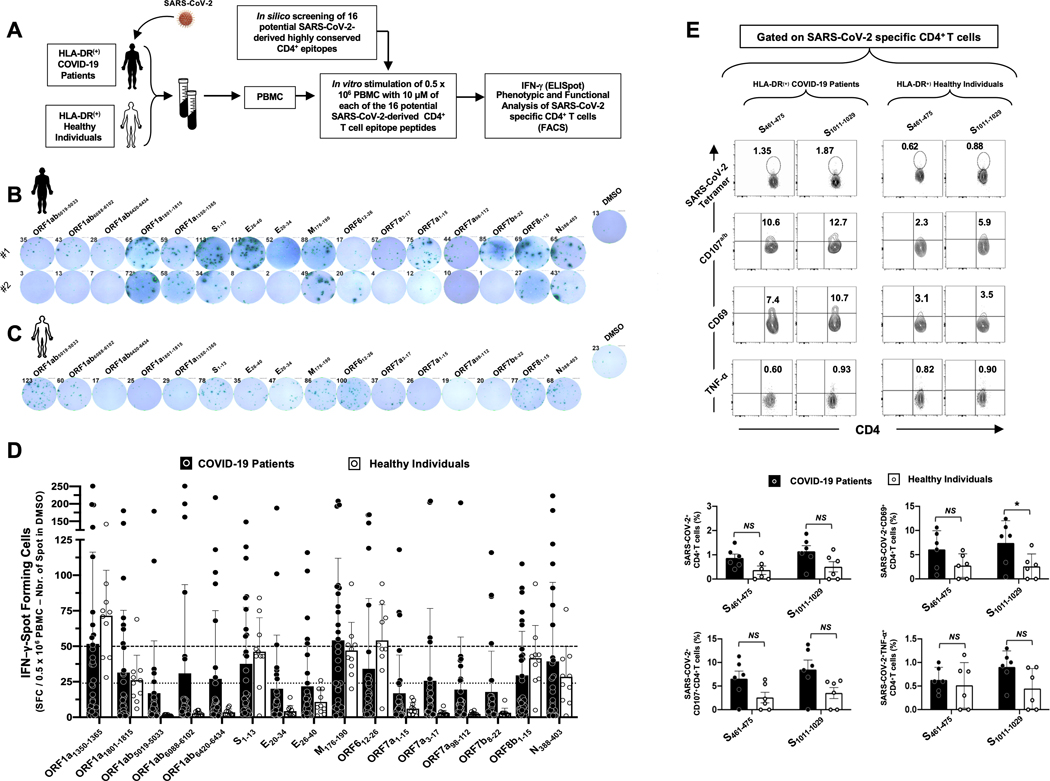
Figure 3: CD8+ T cells specific to highly conserved SARS-CoV-2 epitopes detected in COVID-19 patients and unexposed healthy individuals:
(A) Experimental design: PBMCs from HLA-A*02:01 positive COVID-19 patients (n = 30) (B) and controls unexposed healthy individuals (n = 10) (C) were isolated and stimulated overnight with 10 μM of each of the 27 SARS-CoV-2-derived CD8+ T cell epitopes. The number of IFN-γ-producing cells were quantified using ELISpot assay (B, C and D). Dotted lines represent threshold to evaluate the relative magnitude of the response: a mean SFCs between 25 and 50 correspond to a medium/intermediate response whereas a strong response is defined for a mean SFCs > 50. PBMCs from HLA-A*02:01 positive COVID-19 patients (E) were further stimulated for an additional 5 hours in the presence of mAbs specific to CD107a and CD107b, and Golgi-plug and Golgi-stop. Tetramers specific to Spike epitopes, CD107a/b and CD69 and TNF-α expression were then measured by FACS. Representative FACS plot showing the frequencies of Tetramer+CD8+ T cells, CD107a/b+CD8+ T cells, CD69+CD8+ T cells and TNF-α+CD8+ T cells following priming with a group of 4 Spike CD8+ T cell epitope peptides. Average frequencies of tetramer+CD8+ T cells, CD107a/b+CD8+ T cells, CD69+CD8+ T cells and TNF-α+CD8+ T cells.
Figure 4: CD4+ T cells specific to highly conserved SARS-CoV-2 epitopes detected in COVID-19 patients and unexposed healthy individuals:
(A) Experimental design: PBMCs from HLA-DRB1 positive COVID-19 patients (n = 30) (B) and controls unexposed healthy individuals (n = 10) (C) were isolated and stimulated for 48 hrs. with 10 μM of each of the 16 SARS-CoV-2-derived CD4+ T cell epitopes. The number of IFN-γ-producing cells were quantified using ELISpot assay (B, C and D). Dotted lines represent a threshold to evaluate the relative magnitude of the response: a mean SFCs between 25 and 50 correspond to a medium/intermediate response, whereas a strong response is defined for a mean SFCs > 50. PBMCs from HLA-DRB1-positive COVID-19 patients (E) were further stimulated for an additional 5 hours in the presence of mAbs specific to CD107a and CD107b, and Golgi-plug and Golgi-stop. Tetramers specific to two Spike epitopes, CD107a/b and CD69 and TNF-α expression were then measured by FACS. Representative FACS plot showing the frequencies of Tetramer+CD4+ T cells, CD107a/b+CD4+ T cells, CD69+CD4+ T cells and TNF-α+CD4+ T cells following priming with a group of 2 Spike CD4+ T cell epitope peptides. Average frequencies are shown for tetramer+CD4+ T cells, CD107a/b+CD4+ T cells, CD69+CD4+ T cells and TNF-α+CD4+ T cells.
Blood-derived peripheral blood mononuclear cells (PBMCs) from COVID-19 patients (black, Fig. 3B) and healthy individuals (white, Fig. 3C) were analyzed by ELISpot for frequencies in SARS-CoV-2 epitopes-specific IFN-γ-producing CD8+ T cells. As shown in Figs. 3B and 3D, significant numbers of SARS-CoV-2 epitopes-specific memory CD8+ T cells producing IFN-γ were detected in PBMCs of COVID-19 patients. Out of the 27 highly conserved cross-reactive SARS-CoV-2 CD8+ T cell epitopes (Supplemental Fig. S2) selected for their binding affinity with HLA-A*02:01 molecules (Fig. 2B), strong T cell responses (mean SFCs > 50 per 0.5×106 PBMCs fixed as threshold) were detected in COVID-19 patients against 10 epitopes derived from: (i) structural proteins like Spike (i.e., S958–966, S976–984, S1000–1008 and S1220–1228) or the Envelope proteins (i.e., E26–34) and (ii) non-structural proteins (i.e., ORF1ab1675–1683, ORF1ab2210–2218, ORF1ab6749–6757, ORF63–11, ORF103–11) (Figs. 3B and 3D). In addition, 12 other SARS-CoV-2 CD8+ T cell epitopes from structural of non-structural SARS-CoV-2 proteins induced an intermediate response (with a mean SFCs between 25 and 50 per 0.5×106 PBMCs) in COVID-19 patients: ORF1ab84–92, ORF1ab3013–3021, ORF1ab3183–3191, ORF1ab3732–3740, ORF1ab4283–4291, ORF1ab6419–6427, S2–10, S691–699, E20–28, M52–60, M89–97 and ORF105–13.
Moreover, among the 27 SARS-CoV-2 epitopes, 7 epitopes recalled a strong memory CD8+ T cells response (mean SFCs > 50) from unexposed healthy individuals (i.e., ORF1ab1675–1683, ORF1ab3732–3740, ORF1ab4283–4290, ORF1ab5470–5478, ORF1ab6749–6757, S976–984 and S1000–1008, S1220–1228) and 5 epitopes recalled a memory CD8+ T cells response that was intermediate (ORF1ab6419–6427, S2–10, E26–34, ORF103–11 and ORF105–13) (Figs. 3C and Fig. 3D). However, the unexposed healthy individuals exhibited a different pattern of CD8+ T cell immunodominance as compared to COVID-19 patients. We then compared the epitopes-specificity and function of memory CD8+ T cells in HLA-*A02:01-positive COVID-19 patients and healthy individuals using flow cytometry (Fig. 3E). For a better comparison, a similar FACS gating strategy was applied to PBMCs-derived T cells from both COVID-19 and healthy donors (data not shown). Our COVID-19 patients appeared to have a higher frequency of CD8+ T cells compared to healthy donors (Fig. 3E). Tetramer staining showed that many of SARS-CoV-2 epitope-specific CD8+ T cells are multifunctional producing IFN-γ, TNF-α and expressing CD69 and CD107a/b markers of activation and cytotoxicity in COVID-19 patients (Fig. 3E).
Similar to SARS-CoV-2 memory CD8+ T cells, memory CD4+ T cells specific to several highly conserved SARS-CoV-2 epitopes were detected in both COVID-19-recovered patients and unexposed healthy individuals (Figs. 4B–D). Out of the 16 highly conserved cross-reactive SARS-CoV-2 CD4+ T cell epitopes (Supplemental Fig. S4), strong T cell responses (mean SFCs > 50 per 0.5×106 PBMCs fixed as a threshold) were detected in COVID-19 patients against 2 epitopes, one derived from the structural protein M (M176–190) and one from the non-structural protein ORF1a (ORF1a1350–1365) (Figs. 4B and 4D). Moreover, 6 additional SARS-CoV-2 CD8+ T cell epitopes from non-structural SARS-CoV-2 proteins (i.e., ORF1a1801–1815, ORF1a6088–6102, ORF1a6420–6434, ORF612–26, ORF7a3–17 and ORF8b1–15) and two more epitopes from structural proteins (i.e., S1–13 and N388–403) induced an intermediate CD4+ T cell response (mean SFCs between 25 and 50 per 0.5×106 PBMCs) in COVID-19 patients (Figs. 4B and 4D).
Besides, among the 16 SARS-CoV-2 epitopes, 2 epitopes recalled a strong memory CD4+ T cells response (mean SFCs > 50) from unexposed healthy individuals with no history of COVID-19 (i.e., ORF1a1350–1365 and ORF612–26) (Figs. 4C and 4D). Furthermore, 5 additional epitopes recalled an intermediate CD4+ T cells response in these unexposed healthy individuals (i.e., ORF1a1801–1815, S1–13, M176–190, ORF8b1–15 and N388–403). Unlike for CD8+ T cell responses, the unexposed healthy individuals exhibited a similar pattern of CD4+ T cell immunodominance as compared to COVID-19 patients, with few differences in the magnitude of the responses only. Multifunctional SARS-CoV-2 epitopes-specific CD4+ T cells, expressing CD69, CD107a/b and TNF-α, were detected using specific tetramers in PBMCs of HLA-DR1 positive COVID-19 patients and healthy individuals (Figs. 4E) with a trend showing higher percentage of these cells in COVID-19 patients, although not significantly higher.
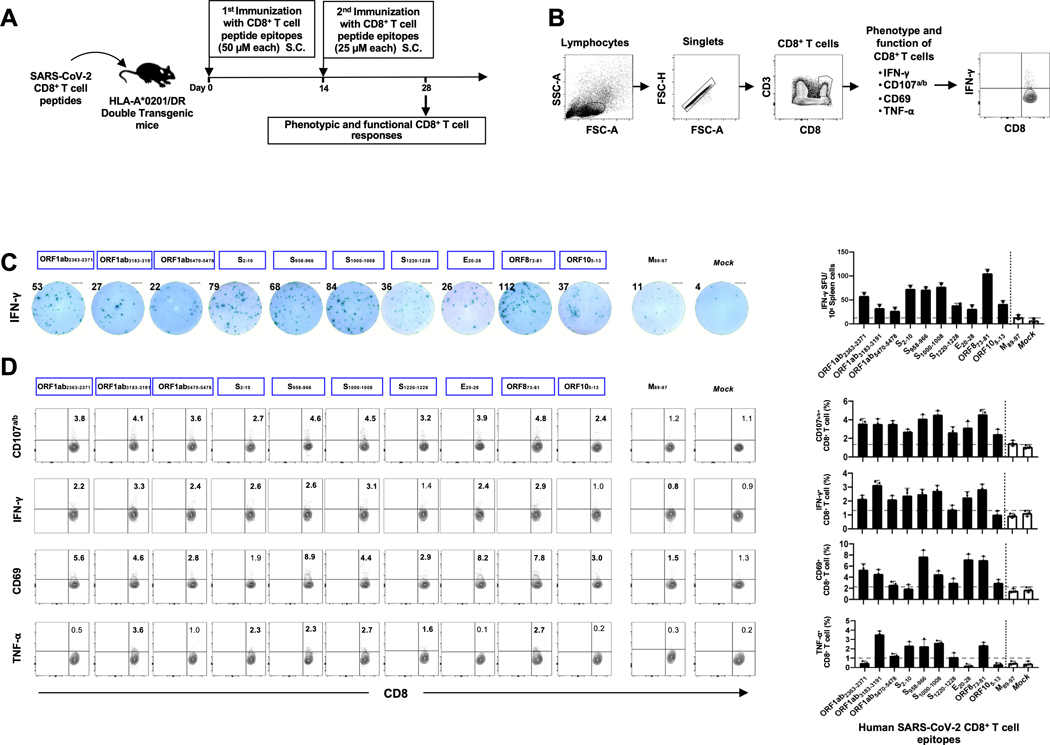
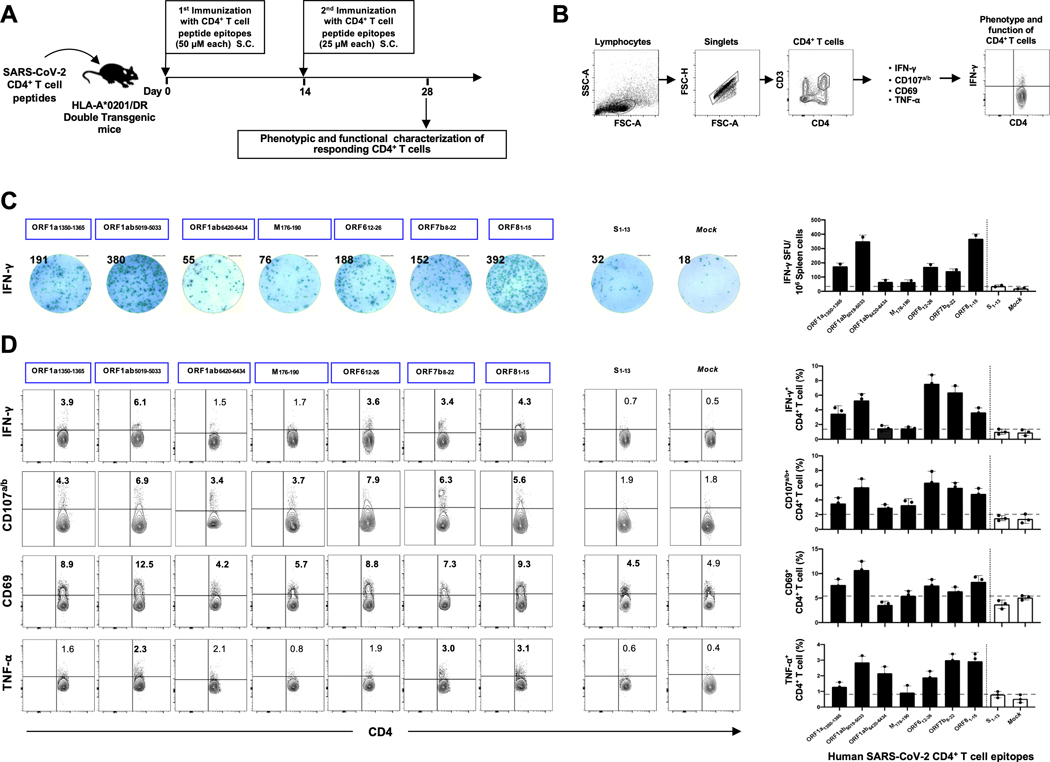
The immunogenicity of the identified SARS-CoV-2 human CD4+ and CD8+ T cell epitopes was assessed in “humanized” HLA-DR1/HLA-A*02:01 double transgenic mice (Figs. 5A and 6A). A mixture of peptides incorporating CD4+ T-cell or CD8+ T-cell epitopes were delivered with CpG and Alum, as shown in Figs. 5A and 6A and detailed in the Materials and Methods. As a negative control, mice received adjuvant alone. The induced SARS-CoV-2 epitope-specific CD4+ and CD8+ T cell responses were determined in the spleen using multiple immunological assays, including IFN-γ ELISpot, FACS surface markers of activation, markers of cytotoxic degranulation and intracellular cytokine staining. The gating strategy used for mice is shown in Figs. 5B and 6B. Two weeks after the second immunization with the mixture of CD8+ T-cell peptides, 10 out of 27 highly conserved SARS-CoV-2 human CD8+ T cell epitope peptides were immunogenic in “humanized” HLA-DR1/HLA-A*02:01 double transgenic mice (Figs. 5C and 5D). The remaining 17 CD8+ T cell epitopes presented moderate/low immunogenicity levels in HLA-DR1/HLA-A*02:01 double transgenic mice. The immunogenic epitopes were derived from both structural Spike protein (S2–10, S958–966, S1000–1008 and S1220–1228) and Envelope protein (E20–28) and from non-structural proteins (i.e., ORF1ab2363–2371, ORF1ab3732–3740, ORF1ab5470–5478, ORF873–81, and ORF105–13). Moreover, 7 out of 16 SARS-CoV-2 peptides induced significant CD4+ T-cell responses in “humanized” HLA-DR1/HLA-A*02:01 double transgenic mice (Figs. 6C and 6D). The immunogenic epitopes were derived from both structural Spike protein (S1–13) and membrane protein (M176–190) and from non-structural proteins (ORF1a1350–1365, ORF1a5019–5033, ORF1a6420–6434, ORF612–26, ORF7b8–22 and ORF8b1–15). The remaining 9 CD4+ T cell epitopes presented moderate/low level of immunogenicity in HLA-DR1/HLA-A*02:01 double transgenic mice.
Figure 5: Immunogenicity of genome-wide identified human SARS-CoV-2 CD8+ T epitopes in HLA-A*02:01/HLA-DRB1 double transgenic mice.
(A) Timeline of immunization and immunological analyses. Eight groups of age-matched HLA-A*02:01 transgenic mice (n = 3) were immunized subcutaneously, on days 0 and 14, with a mixture of four SARS-CoV-2-derived human CD8+ T cell peptide epitopes mixed with PADRE CD4+ T helper epitope, delivered in alum and CpG1826 adjuvants. As a negative control, mice received adjuvants alone (mock-immunized). (B) Gating strategy used to characterize spleen-derived CD8+ T cells. Lymphocytes were identified by a low forward scatter (FSC) and low side scatter (SSC) gate. Singlets were selected by plotting forward scatter area (FSC-A) vs. forward scatter height (FSC-H). CD8 positive cells were then gated by the expression of CD8 and CD3 markers. (C) Representative ELISpot images (left panel) and average frequencies (right panel) of IFN-γ-producing cell spots from splenocytes (106 cells/well) stimulated for 48 hours with 10 μM of 10 immunodominant CD8+ T cell peptides and 1 subdominant CD8+ T cell peptide out of the total pool of 27 CD8+ T cell peptides derived from SARS-CoV-2 structural and non-structural proteins. The number on the top of each ELISpot image represents the number of IFN-γ-producing spot forming T cells (SFC) per one million splenocytes. (D) Representative FACS plot (left panel) and average frequencies (right panel) of IFN-γ and TNF-α production by, and CD107a/b and CD69 expression on 10 immunodominant CD8+ T cell peptides and 1 subdominant CD8+ T cell peptide out of the total pool of 27 CD8+ T cell peptides derived from SARS-CoV-2 structural and non-structural proteins determined by FACS. Numbers indicate frequencies of IFN-γ+CD8+ T cells, CD107+CD8+ T cells, CD69+CD8+ T cells and TNF-α+CD8+ T cells, detected in 3 immunized mice.
Figure 6: Immunogenicity of genome-wide identified human SARS-CoV-2 CD4+ T epitopes in HLA-A*02:01/HLA-DRB1 double transgenic mice.
(A) Timeline of immunization and immunological analyses. Four groups of age-matched HLA-DRB1 transgenic mice (n = 3) were immunized subcutaneously, on days 0 and 14, with a mixture of four SARS-CoV-2-derived human CD4+ T cell peptide epitopes delivered in alum and CpG1826 adjuvants. As a negative control, mice received adjuvants alone (mock-immunized). (B) Gating strategy used to characterize spleen-derived CD4+ T cells. CD4 positive cells were gated by the CD4 and CD3 expression markers. (C) Representative ELISpot images (left panel) and average frequencies (right panel) of IFN-γ-producing cell spots from splenocytes (106 cells/well) stimulated for 48 hours with 10 μM of 7 immunodominant CD4+ T cell peptides and 1 subdominant CD4+ T cell peptide out of the total pool of 16 CD4+ T cell peptides derived from SARS-CoV-2 structural and non-structural proteins. The number of IFN-γ-producing spot forming T cells (SFC) per one million of total cells is presented on the top of each ELISpot image. (D) Representative FACS plot (left panel) and average frequencies (right panel) show IFN-γ and TNF-α-production by, and CD107a/b and CD69 expression on 7 immunodominant CD4+ T cell peptides and 1 subdominant CD4+ T cell peptide out of the total pool of 16 CD4+ T cell peptides derived from SARS-CoV-2 determined by FACS. The numbers indicate percentages of IFN-γ+CD4+ T cells, CD107+CD4+ T cells, CD69+CD4+ T cells and TNF- α+CD4+ T cells detected in 3 immunized mice.
Altogether, these results indicate that pre-existing memory CD4+ T and CD8+ T cells specific to both structural and non-structural protein antigens and epitopes are present in COVID-19 patients and unexposed healthy individuals. While SARS-CoV-2-specific CD4+ and CD8+ T cells in COVID-19 patients and healthy donors target epitopes from the whole virus proteome, most T cell epitopes are concentrated in the non-structural proteins, with ORF1a/b being the most targeted antigens. These memory T cells recognized highly conserved SARS-CoV-2 epitopes that cross-react with the human and animal Coronaviruses. It is likely that infection with a “common cold” Coronavirus and/or human exposition with animal and pet related coronaviruses induced long-lasting memory CD4+ and CD8+ T cells specific to the structural and non-structural SARS-CoV-2 epitopes in healthy unexposed individuals. Heterologous immunity and heterologous immunopathology orchestrated by these cross-reactive epitope-specific memory CD4+ and CD8+ T cells, following previous multiple exposures to “common cold” Coronaviruses, may have shaped protection versus susceptibility to SARS-CoV-2 infection and disease, with a yet-to-be determined mechanism(s).
5. Identification of B-cell epitopes from SARS-CoV-2 Spike protein that are highly conserved between human and animal Coronaviruses, that are antigenic in humans and immunogenic in “humanized” HLA transgenic mice:
We next predicted potential linear B-cell (antibody) epitopes on Spike protein sequence of the first SARS-CoV-2-Wuhan-Hu-1 strain (NCBI GenBank accession number MN908947.3) using BepiPred 2.0, with a cutoff of 0.55 (corresponding to a specificity greater than 0.81 and sensitivity below 0.30) and considering sequences having more than 5 amino acid residues (67). This stringent screening process initially resulted in the identification of 28 linear B-cell epitopes (Supplemental Table S3). From this pool of 28 potential epitopes, we later selected 15 B-cell epitopes, (19 to 62 amino acids in length), based on: (i) their sequences being highly conserved between SARS-CoV-2, the main 4 major “common cold” Coronaviruses (CoV-OC43 (KF923903), CoV-229E (KY983587), CoV-HKU1 (AY884001), and CoV-NL63 (NC_005831)) (68), and the SARS-like SL-CoVs that are isolated from bats, civet cats, pangolins and camels; and (ii) the probability of exposure each linear epitope to the surface of infected target cells (Supplemental Fig. S6). The Spike epitope sequences highlighted in blue indicate a high degree of homology among the currently circulating 81,963 SARS-CoV-2 strains and at least a 50% conservancy among two or more human SARS-CoV strains from previous outbreaks, and the SL-CoV strains isolated from bats, civet cats, pangolins and camels (Supplemental Fig. S6). Two of the 15 B-cell epitopes namely S369–393, and S440–501 overlap with the Spike’s receptor binding domain (RBD) region that bind to the ACE2 receptor (designated as RBD-1 and RBD-2 in Supplemental Fig. S7A). Higher interaction similarity scores were observed for RBD-derived epitopes S369–393 and S471–501 when molecular docking was performed against the ACE2 receptor (Supplemental Fig. S7B). Upon screening for the glycosylation regions, we observed B-cell epitopes S13–37, S59–81, S329–363, S601–640, S1133–1172 with Asparagines predicted to be N-glycosylated. In contrast, B-cell epitopes S516–536, S524–598, S802–819 were observed to be the O-glycosylated. The remaining B-cell epitopes S287–317, S304–322, S369–393, S404–501, S440–501, S672–690, and S888–909 were found to possess no glycosylation.
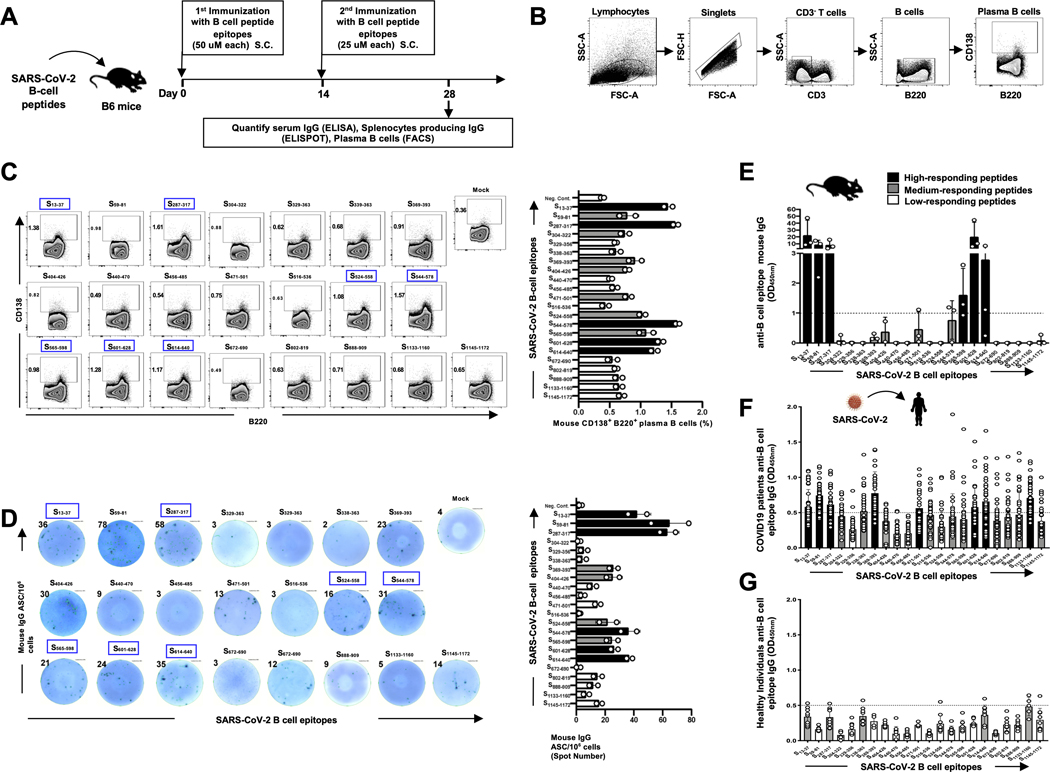
We later determined the ability of each of the 15 B-cell epitopes selected from the Spike protein, that showed a high conservancy between human and animal Coronaviruses, to induce SARS-CoV-2 epitope-specific antibody-producing plasma B cells and IgG antibodies in B6 mice (Fig. 7). Synthetic peptides corresponding to each linear B cell epitope were produced. Since 4 epitopes were too long to synthetize (e.g., 62 amino acids), they were divided into 2 or 3 short fragments resulting in a total of 22 B-cell epitope peptides (Supplemental Table S3). As illustrated in Fig. 7A, groups of five B6 mice each received two subcutaneous (s.c.) injections with mixtures of 3 to 4 B-cell epitope peptides, mixed with CpG and Alum adjuvants. Negative control mice received adjuvant alone, without Ags. The frequency of antibody-producing plasma B cells and the level of IgG antibodies specific to each SARS-CoV-2 B cell epitope were determined in the spleen and in the serum using FACS staining of CD138 and B220 surface markers and IgG-ELISpot and ELISA assays, respectively. The gating strategy used to determine the frequencies of plasma B-cells in the spleen is shown in Fig. 7B. Out of the 22 Spike B-cell epitopes, 7 epitopes (S13–37, S287–317, S524–558, S544–578, S565–598, S601–628, and S614–640) induced high frequencies of CD138+B220+ plasma B cells in the spleen of B6 mice (Fig. 7C). The IgG ELISpot assay confirmed that 7 out of the 22 Spike B-cell epitopes induced significant numbers of IgG-producing B cells in the spleen (Fig. 7D). Moreover, significant amounts of IgG were detected in the serum of the immunized B6 mice. These IgG antibodies were specific to 6 out of the 22 Spike B-cell peptide epitopes (S13–37, S59–81, S287–317, S565–598, S601–628, and S614–640) (Fig. 7E). As expected, non-immunized animals or those that received adjuvant alone did not develop detectable IgG responses. Of these 6 highly immunogenic B cell peptides, 5 peptides (S13–37, S59–81, S287–317, S601–628, and S614–640) were highly antigenic as they were recognized by serum IgG from COVID-19 patients, confirming the presence of at least one native linear B cell epitope in each peptide (Fig. 7F). In summary, we identified five highly conserved immunogenic and antigenic human B-cell target epitopes from the Spike SARS-CoV-2 virus that recall IgG antibodies from COVID-19 patients. This study further discovered five highly conserved B-cell epitopes from SARS-CoV-2: S13–37, S287–317, S338–363, S614–640, and S1133–1160 that are recognized by IgG antibodies from healthy individuals who were never exposed to COVID-19, suggesting B-cell epitopes cross-reactivity to other human Coronaviruses (Fig. 7G).
Figure 7: IgG antibodies specific to SARS-CoV-2 Spike protein-derived B-cell epitopes in immunized B6 mice and in convalescent COVID-19 patients:
(A) Timeline of immunization and immunological analyses. A total of 22 SARS-CoV-2 derived B-cell epitope peptides selected from SARS-CoV-2 Spike protein and tested in B6 mice were able to induce antibody responses. Four groups of age-matched B6 mice (n = 3) were immunized subcutaneously, on days 0 and 14, with a mixture of 4 or 5 SARS-CoV-2 derived B-cell peptide epitopes emulsified in alum and CpG1826 adjuvants. Alum/CpG1826 adjuvants alone were used as negative controls (mock-immunized). (B and C) The frequencies of IgG-producing CD3(−)CD138(+)B220(+) plasma B cells were determined in the spleen of immunized mice by flow cytometry. (B) The gating strategy was as follows: Lymphocytes were identified by a low forward scatter (FSC) and low side scatter (SSC) gate. Singlets were selected by plotting forward scatter area (FSC-A) versus forward scatter height (FSC-H). B cells were then gated by the expression of CD3(−) and B220(+) cells and CD138 expression on plasma B cells determined. (C) Representative FACS plot (left panels) and average frequencies (right panel) of plasma B cells detected in spleen of immunized mice. The percentages of plasma CD138(−)B220(+)B cells is indicated on the top left of each dot plot. (D) SARS-CoV-2 derived B-cell epitopes-specific IgG responses were quantified in immune serum, 14 days post-second immunization (i.e. day 28), by ELISpot (Number of IgG(+)Spots). Representative ELISpot images (left panels) and average frequencies (right panel) of anti-peptide specific IgG-producing B cell spots (1×106 splenocytes/well) following 4 days in vitro B cell polyclonal stimulation with mouse Poly-S (Immunospot). The top/left of each ELISpot image shows the number of IgG-producing B cells per half a million cells. ELISA plates were coated with each individual immunizing peptide. The B-cell epitopes-specific IgG concentrations (μg/mL) measured by ELISA in: (E) Levels of IgG detected in peptide-immunized B6 mice, after subtraction of the background measured from mock-vaccinated mice. The dashed horizontal line indicates the limit of detection; and in (F) Level of IgG specific to each of the 22 Spike peptides detected SARS-CoV-2 infected patients (n=40), after subtraction of the background measured from healthy non-exposed individuals, as shown in (G) (n=10). Black bars and gray bars show high and medium immunogenic B cell peptides, respectively. The dashed horizontal line indicates the limit of detection.
DISCUSSION
While the current COVID-19 pandemic will likely be overcome through the implementation of physical distancing, barriers together with a mass vaccination, it is indispensable that a safe and effective pre-emptive vaccine be developed, and in place ready to protect, against another inevitable COVID pandemic that will emerge in the years to come.
Towards this goal of developing a multi-epitope pre-emptive pan-Coronavirus human vaccine, we identified several cross-reactive human B and T cell epitopes of SARS-CoV-2 that are highly conserved among the human SARS-CoVs and animal SL-CoVs. While antiviral SARS-CoV-2-specific antibodies and CD4+ and CD8+ T cell responses appear crucial in protecting asymptomatic patients and convalescent patients, very little information exists with regards to the repertoire of targeted SARS-CoV-2 B and T cell epitopes that are common within a substantial group of human and animal Coronaviruses (58, 69, 70). In agreement with our results, 4 out of the 27 CD8+ T cell epitopes reported in this study, have been recently reported to be cross-reactive between SARS-CoV and SARS-CoV-2: ORF1ab2363–2371, ORF1ab3013–3021, S958–966 and S1220–1228 (58, 70). Similarly, B cell epitope S287–317, has been reported as cross-reactive between SARS-CoV and SARS-CoV-2(58, 70). However, to the best of our knowledge, none of the 16 CD4+ T cell epitopes identified herein have been reported previously. The highly conserved human B and T cell epitopes reported in this study have massive implications for the development of an universal pre-emptive pan-Coronavirus vaccine to induce (or to boost) neutralizing antibodies (Abs), CD4+ T helpers (Th1), and antiviral CD8+ cytotoxic T-cells (CTL) (21, 71, 72). Some of our identified epitopes are similar to those recently reported by Grifoni, et al, (58, 70) while other epitopes have never been reported. Moreover, in agreement with recent reports (58, 70, 73), our study revealed a high degree of similarity among SARS-CoV-2, SARS-CoV and bat-SL-CoVs epitope sequences, but not the MERS-CoV epitope sequences. In the present study we have identified B-cell epitopes S13–37, S59–81, S329–363, S601–640, S1133–1172 with N-glycosylated regions, and S516–536, S524–598, S802–819 with O-glycosylated regions. Extensive glycosylation has been observed in CoV’s surface glycoproteins, representing the most extensive known class I viral fusion proteins. SARS spike glycoprotein is known to encode 69 N-linked glycan sequons per trimeric spike with SARS-CoV-2 containing 66 sites (74). These modifications may mask immunogenic B cell epitopes from the host humoral immune system by occluding them with host-derived glycans. However, some studies underscore the importance of glycosylation in the lack of immunogenicity and viral immune evasion. Watanabe et al. have reported that extensive N-linked glycan modifications of SARS and MERS CoV surface glycoproteins do not constitute an effective shield compared to glycan shields of certain other viruses, which is reflected by the overall structure, density, and oligomannose abundance across the corresponding trimeric glycoproteins 68.
Whether the identified epitopes will contribute to protection is beyond the scope of the present study. Unlike most Coronavirus subunit vaccines (75–77), our multi-epitope Coronavirus vaccine (e.g., the pan-Coronavirus candidate #1 illustrated in Supplemental Fig. S8) incorporates multiple human asymptomatic B and CD4+/CD8+ T cell epitopes that are selected carefully from the whole genome of SARS-CoV-2 for being recognized by antibodies and CD4+/CD8+ T cells from asymptomatic and convalescent patients that are “naturally protected” from COVID-19. The present study employed a combinatorial approach for designing an all-in-one multi-epitope pan-Coronavirus vaccine candidate (Supplemental Fig. S8) by applying highly conserved genome-wide human B- and T-cell epitopes from 12 genome derived antigenic proteins of SARS-CoV-2. The present study focused on HLA-A*02:01-restricted epitopes represented by more than 50% of the human population. However, epitopes restricted to other HLA-A, HLA-B, and HLA-C haplotypes, including the forecasted population coverage of the chosen T cell epitope ensemble (combined HLA class I), are expected to cover 99.8% of the global population regardless of race and ethnicity. In addition, for a wider vaccine coverage (i.e., close to 99%), our multi-epitope pan-Coronavirus vaccine platform would be easily adapted to include CD8+ T cell epitopes for other HLA supertypes that are distributed in the various human populations. The polymorphic HLA molecules can be clustered into a handful of HLA-A supertypes that bind largely overlapping peptide repertoires (78). Moreover, such a multi-epitope vaccine would be easily adapted to exclude undesirable epitopes that are restricted to HLA-B*44 and HLA-C*01 alleles which appear to correlate with SARS-CoV-2 virus spreading across certain countries (79) and HLA-B*35 allele which appear to be associated with severe pneumonia developed by SARS-CoV-2 in young patients (80).
We do not exclude that the highly conserved B-cell, CD4+ and CD8+ T cell epitopes identified in from bat’s Coronavirus variants will mutate, following recombination that often occurs for zoonotic events before an animal SL-CoV spills over into humans. In this context, our pre-emptive multi-epitope pan-Coronavirus vaccine is highly adaptable to newly mutated Coronavirus strains. If a Coronavirus epitope mutates, that single epitope can be easily adjusted and replaced in the multi-epitope vaccine (81). In lieu of this, we have screened all of our candidate epitopes against SARS-CoV-2 variants, which have been evidenced with increased transmissibility, including B.1.1.7 variant emerging from the UK (variant 20I/501Y.V1), B.1.351 variant emerging from South Africa (variant 20H/501Y.V2), B.1.1.28 variant emerging from Brazil (P.1 variant 20J/501Y.V3), CAL.20C variant observed in California (Supplemental Fig. S9). We found 100% conservancy for 15 out of 16 CD4+ T cell epitopes against B.1.1.7 (United Kingdom), B.1.351 (South Africa), B.1.1.28 (Brazil) variants. One mutation in S13I region that was found in the CAL.20C variant is observed in one of our CD4+ T cell epitopes S1–13 (MFVFLVLLPLVSS), whereas the remaining 15 CD4+ T cell epitopes showed 100% conservancy CAL.20C variant. Notably, all our 27 CD8+ T cell epitopes showed conservancy against highly transmissible South Africa, Brazil, California (CAL.20C) variants. However, one region specific to the non-synonymous mutation S982A from the UK variant was observed in our CD8+ T cell epitope S976–984 (VLNDILSRL). The remaining 26 CD8+ T cell epitopes showed 100% conservancy with the B.1.1.7 (United Kingdom) variant. In comparison, two of the screened B-cell epitopes (S59–81, S601–640) belonged to regions specific to South African variant B.1.351 (20H/501Y.V2). More so, two B-cell epitopes (S404–426, S440–501) represent the regions specific to South African variant B.1.351 (20H/501Y.V2), Brazilian variant B.1.1.28 (P.1 variant 20J/501Y.V3), and California variant (CAL.20C). Two B-cell epitopes S524–598 and S672–690 represent regions specific to non-synonymous spike protein-specific mutations A570D and P681H found in the B1.1.7 strain from the UK. This emphasizes that our pre-emptive multi-epitope pan-Coronavirus vaccine strategy could be easily adapted to any variant as well as to any new zoonotic bat SL-CoVs that may spill over in the future into humans in the future. This high adaptability is expected to speed up the implementation of a future pre-emptive multi-epitope vaccine before a local outbreak spreads and transforms into a global pandemic.
It is inevitable that future COVID-like outbreaks, caused by yet another spillover of a bat SL-CoV, could lead to other COVID-like pandemics with global health, social and economic disasters in the years to come. However, because it is almost impossible to predict which viral strain might cause the next Coronavirus pandemic, it is urgent to develop a pan-Coronavirus vaccine that targets a wide range of human and animal Coronavirus strains. Unlike conventional monovalent vaccines made from epitopes selected from a single virus strain, a pre-emptive multi-epitope pan-Coronavirus vaccine (Supplemental Fig. S8) that includes several highly conserved human B-, CD4+ and CD8+ T cell epitopes identified from the entire genome sequences of human SARS-CoVs that cross-react and shared with bat and pangolin SL-CoVs (18, 82–85). The current ongoing collaborative research efforts should not only focus on developing a vaccine for COVID-19, but should also be oriented towards developing pre-emptive pan-Coronavirus vaccines. Such a proactive vaccine strategy would help fight and contain future local outbreaks and epicenters of highly contagious and deadly zoonotic Coronaviruses globally before becoming the next deadly pandemic worldwide (21, 72). Moreover, because it is impossible to predict the time and location of the next deadly global pandemic, it is essential to have ready, at least pre-clinically, several pan-Coronavirus vaccine candidates that would be quickly implemented in a clinical trial against a substantial group of Coronaviruses before an outbreak spreads into a global pandemic(s).
In the present study, CD4+ and CD8+ T cells specific to highly conserved SARS-CoV2 epitopes were detected in healthy adults, recruited between 2014 and 2018, who have never been exposed to the SARS-CoV-2 virus. These findings suggest cross-reactive T cells between current SARS-CoV-2 and previous circulating “common cold” Coronaviruses, as confirmed by recent reports (70, 86, 87). However, since it is unknown whether the healthy adults used in this study were indeed exposed to any “common cold” Coronaviruses, such an assertion may not be conclusive. Among the many circulating “common cold” Coronaviruses known to infect humans, four serotypes that cause severe respiratory infections are highly seasonal: CoV-OC43, CoV-229E, CoV-HKU1, and CoV-NL63 (68) and appear to have a similar transmission potential to influenza A (H3N2). The seasonality of these “common cold” Coronaviruses is predictable as their outbreaks often emerged in December, peaked in January/February, and began to decrease in March of every year (68).
The human SARS-CoV-2 CD4+ and CD8+ T cell epitopes identified in this study are highly conserved between 81,963 strains of SARS-CoV-2 and CoV-OC43, CoV-229E, CoV-HKU1, and CoV-NL63 (Supplemental Figs. S2 and S4). Whether these apparent cross-reactive CD4+ and CD8+ T cells play a protective or a harmful role or an entirely negligible role in SARS-CoV-2 infection and disease remains to be determined (86, 87). Nevertheless, since “common cold” Coronavirus infections are frequent in children, it will be interesting to determine whether children who appeared more resistant to COVID-19 compared to adults will have robust antiviral memory T cell responses to some of the common SARS-CoV-2 epitopes identified in this study. A stronger CD4+ and CD8+ T cell responses to common Coronavirus epitopes in children would shed some light on the unique situation currently seen in COVID-19 where immune children tend to be more resistant to SARS-CoV-2 infection and disease, as compared to more susceptible adults (88, 89). Such a result would also imply that a pan-Coronavirus vaccine incorporating these cross-reactive highly conserved SARS-CoV-2 human CD4+ and CD8+ T cell epitopes would boost protective T cell immunity that would have been previously induced by a “common cold” Coronavirus. This would protect not only from seasonal circulating “common cold” Coronaviruses, but also from SARS-CoV-2 infection and disease.
Even though the highly conserved Coronavirus human CD4+ and CD8+ T cell epitopes identified in this report can be enlightening for a pan-Coronavirus vaccine, humans are not immunologically naive, and they often have memory CD4+ and CD8+ T cell populations that can cross-react with, and respond to, other infectious agents, a phenomenon termed heterologous immunity (90). Therefore, we cannot exclude that some SARS-CoV-2-specific CD4+ and CD8+ T cell epitopes identified in this study are cross-reactive with other viral pathogens-derived epitopes, such as epitopes from circulating seasonal influenza or “common cold” Coronaviruses (91, 92). This may explain, in part, the high proportion of asymptomatic infections with SARS-CoV-2 in the current pandemic. The latter is supported by a recent elegant study that detected SARS-CoV-2-reactive CD8+ and CD4+ T cells in healthy individuals that were never exposed to SARS-CoV-2 (70). SARS-CoV-2-specific, but cross-reactive, CD4+ and CD8+ T cells can become activated and modulate the immune responses and clinical outcome of subsequent heterologous SARS-CoV-2 infections. Therefore, T cell cross-reactivity may be crucial in protective heterologous immunity instead of damaging heterologous immunopathology, as has been reported in other systems (93). To confirm SARS-CoV-2 heterologous CD4+ and CD8+ T cell epitopes that may potentially cross-react with other pathogenic (non-Coronaviruses) epitopes, we are currently comparing the CD4+ and CD8+ T-cell response to those highly conserved SARS-CoV-2 epitopes identified using humans CD4+ and CD8+ T-cell responses to those of “pathogen-free” SARS-CoV-2-infected transgenic mice.
In conclusion, we report here several previously unknown human “universal” B, and CD4+ and CD8+ T cell target epitopes identified from the whole SARS-CoV-2 genome. These epitopes are highly conserved and common between SARS-CoV-2 Wuhan strain and: (1) seven circulating “common cold” human Coronaviruses that caused previous human SARS and MERS outbreaks (72); (2) 81,963 strains of human SARS-CoV-2 that now circulate in six continents; (3) several bat-derived SL-CoV strains (13, 14); and (4) several SL-CoV strains isolated from pangolins (94). The findings from this report pave the way for the development of “pre-emptive” multi-epitope pan-Coronavirus vaccine candidates that would target not only the current human COVID-19 outbreak, but also possible future Coronavirus outbreaks that might come from a bat-derived SL-CoV strain that would spill over again into humans.
Supplementary Material
KEY POINTS.
Highly conserved B and T cell epitopes identified in human and animals Coronaviruses
Cross-reactive Coronavirus epitopes recognized by COVID-19 and healthy patients
Conserved SARS-CoV-2 epitopes are immunogenic in “humanized” HLA transgenic mice
ACKNOWLEDGMENTS
The authors would like to thank Dr. Dale Long from the NIH Tetramer Facility (Emory University, Atlanta, GA) for providing the Tetramers used in this study. We thank UC Irvine Center for Clinical Research (CCR) and Institute for Clinical & Translational Science (ICTS) for providing human blood samples used in this study. A special thanks to Dr. Delia F. Tifrea for her continuous efforts and dedication in providing COVID-19 samples that are crucial for this clinical research. We also thank those who contributed directly or indirectly to this COVID-19 vaccine project: Dr. Steven A. Goldstein, Dr. Michael J. Stamos, Dr. Suzanne B. Sandmeyer, Jim Mazzo, Dr. Daniela Bota, Dr. Beverly L. Alger, Dr. Dan Forthal, Dr. Tahseen Muzaffar, Dr. Ilhem Messaoudi, Anju Subba, Janice Briggs, Marge Brannon, Beverley Alberola, Jessica Sheldon, Rosie Magallon and Andria Pontello.
This work is supported by the Fast-Grant PR12501 from Emergent Ventures, by a Gavin Herbert Eye Institute internal grant, by Public Health Service Research grants AI158060, AI150091, AI143348, AI147499, AI143326, AI138764, AI124911 and AI110902 from the National Institutes of Allergy and Infectious Diseases (NIAID) to LBM.
Footnotes
Declaration of Interest: The University of California Irvine has filed a patent application on the results reported in this manuscript.
REFERENCES
- 1.Carroll D, Watson B, Togami E, Daszak P, Mazet JA, Chrisman CJ, Rubin EM, Wolfe N, Morel CM, Gao GF, Burci GL, Fukuda K, Auewarakul P, and Tomori O. 2018. Building a global atlas of zoonotic viruses. Bull World Health Organ 96: 292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen T, Murray KA, Zambrana-Torrelio C, Morse SS, Rondinini C, Di Marco M, Breit N, Olival KJ, and Daszak P. 2017. Global hotspots and correlates of emerging zoonotic diseases. Nat Commun 8: 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valitutto MT, Aung O, Tun KYN, Vodzak ME, Zimmerman D, Yu JH, Win YT, Maw MT, Thein WZ, Win HH, Dhanota J, Ontiveros V, Smith B, Tremeau-Brevard A, Goldstein T, Johnson CK, Murray S, and Mazet J. 2020. Detection of novel coronaviruses in bats in Myanmar. PloS one 15: e0230802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijaykrishna D, Smith GJ, Zhang JX, Peiris JS, Chen H, and Guan Y. 2007. Evolutionary insights into the ecology of coronaviruses. J Virol 81: 4012–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masters PS 2006. The molecular biology of coronaviruses. Adv Virus Res 66: 193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, and Poon LL 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302: 276–278. [DOI] [PubMed] [Google Scholar]
- 7.Pyrc K, Jebbink MF, Berkhout B, and van der Hoek L. 2004. Genome structure and transcriptional regulation of human coronavirus NL63. Virol J 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farsani SM, Dijkman R, Jebbink MF, Goossens H, Ieven M, Deijs M, Molenkamp R, and van der Hoek L. 2012. The first complete genome sequences of clinical isolates of human coronavirus 229E. Virus Genes 45: 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, and Talbot PJ 2004. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J Virol 78: 8824–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu G, Wang Q, and Gao GF 2015. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol 23: 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wurzer WJ, Obojes K, and Vlasak R. 2002. The sialate-4-O-acetylesterases of coronaviruses related to mouse hepatitis virus: a proposal to reorganize group 2 Coronaviridae. J Gen Virol 83: 395–402. [DOI] [PubMed] [Google Scholar]
- 12.Agnihothram S, Yount BL Jr., Donaldson EF, Huynh J, Menachery VD, Gralinski LE, Graham RL, Becker MM, Tomar S, Scobey TD, Osswald HL, Whitmore A, Gopal R, Ghosh AK, Mesecar A, Zambon M, Heise M, Denison MR, and Baric RS 2014. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. mBio 5: e00047–00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menachery VD, Yount BL Jr., Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, Swanstrom J, Sheahan TP, Pickles RJ, Corti D, Randell SH, Lanzavecchia A, Marasco WA, and Baric RS 2016. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 113: 3048–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menachery VD, Yount BL Jr., Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, and Baric RS 2016. Corrigendum: A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 22: 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menachery VD, Yount BL Jr., Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, and Baric RS 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21: 1508–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Zheng X, Tong Q, Li W, Wang B, Sutter K, Trilling M, Lu M, Dittmer U, and Yang D. 2020. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol 92: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menachery VD, Graham RL, and Baric RS 2017. Jumping species-a mechanism for coronavirus persistence and survival. Current opinion in virology 23: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, and Shi ZL 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan Y, Shang J, Graham R, Baric RS, and Li F. 2020. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layne SP, Hyman JM, Morens DM, and Taubenberger JK 2020. New coronavirus outbreak: Framing questions for pandemic prevention. Science translational medicine 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lun ZR, and Qu LH 2004. Animal-to-human SARS-associated coronavirus transmission? Emerg Infect Dis 10: 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowell G, and Mizumoto K. 2020. The COVID-19 pandemic in the USA: what might we expect? Lancet 395: 1093–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decru B, Van Elslande J, Weemaes M, Houben E, Empsen I, Andre E, Van Ranst M, Lagrou K, and Vermeersch P. 2020. Comparison of the diagnostic performance with whole blood and plasma of four rapid antibody tests for SARS-CoV-2. Clin Chem Lab Med 58: e197–e199. [DOI] [PubMed] [Google Scholar]
- 24.An P, Song P, Wang Y, and Liu B. 2020. Asymptomatic Patients with Novel Coronavirus Disease (COVID-19). Balkan Med J 37: 229–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oran DP, and Topol EJ 2020. Prevalence of Asymptomatic SARS-CoV-2 Infection : A Narrative Review. Ann Intern Med 173: 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breslin N, Baptiste C, Gyamfi-Bannerman C, Miller R, Martinez R, Bernstein K, Ring L, Landau R, Purisch S, Friedman AM, Fuchs K, Sutton D, Andrikopoulou M, Rupley D, Sheen JJ, Aubey J, Zork N, Moroz L, Mourad M, Wapner R, Simpson LL, D’Alton ME, and Goffman D. 2020. COVID-19 infection among asymptomatic and symptomatic pregnant women: Two weeks of confirmed presentations to an affiliated pair of New York City hospitals. Am J Obstet Gynecol MFM: 100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An P, Song P, Wang Y, and Liu B. 2020. Asymptomatic Patients with Novel Coronavirus Disease (COVID-19). Balkan Med J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DK, Agnihothram S, Baric RS, David CS, and Perlman S. 2016. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 44: 1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Hou J, Jiang X, Ma S, Meng M, Wang B, Zhang M, Zhang M, Tang X, Zhang F, Wan T, Li N, Yu Y, Hu H, Yang R, He W, Wang X, and Cao X. 2005. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J Immunol 175: 591–598. [DOI] [PubMed] [Google Scholar]
- 30.Rajendran K, Krishnasamy N, Rangarajan J, Rathinam J, Natarajan M, and Ramachandran A. 2020. Convalescent plasma transfusion for the treatment of COVID-19: Systematic review. J Med Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, and Corti D. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. [DOI] [PubMed] [Google Scholar]
- 32.Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Ge J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin CF, Chen F, and Dong C. 2020. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee CY, Lin RTP, Renia L, and Ng LFP 2020. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front Immunol 11: 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Bruggen MC, O’Mahony L, Gao Y, Nadeau K, and Akdis CA 2020. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Ge J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin CF, Chen F, and Dong C. 2020. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 52: 971–977 e973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, and Hlh UK Across Speciality Collaboration. 2020. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395: 1033–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaninov N. 2020. In the eye of the COVID-19 cytokine storm. Nat Rev Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J, Xia P, Zhou Y, Liu Z, Zhou X, Wang J, Li T, Yan X, Chen L, Zhang S, Qin Y, and Li X. 2020. Potential effect of blood purification therapy in reducing cytokine storm as a late complication of critically ill COVID-19. Clin Immunol 214: 108408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson LA, Canna SW, Schulert GS, Volpi S, Lee PY, Kernan KF, Caricchio R, Mahmud S, Hazen MM, Halyabar O, Hoyt KJ, Han J, Grom AA, Gattorno M, Ravelli A, de Benedetti F, Behrens EM, Cron RQ, and Nigrovic PA 2020. On the alert for cytokine storm: Immunopathology in COVID-19. Arthritis & rheumatology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh KB, Teijaro JR, Brock LG, Fremgen DM, Collins PL, Rosen H, and Oldstone MB 2014. Animal model of respiratory syncytial virus: CD8+ T cells cause a cytokine storm that is chemically tractable by sphingosine-1-phosphate 1 receptor agonist therapy. J Virol 88: 6281–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matheu MP, Teijaro JR, Walsh KB, Greenberg ML, Marsolais D, Parker I, Rosen H, Oldstone MB, and Cahalan MD 2013. Three phases of CD8 T cell response in the lung following H1N1 influenza infection and sphingosine 1 phosphate agonist therapy. PloS one 8: e58033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh KB, Teijaro JR, Wilker PR, Jatzek A, Fremgen DM, Das SC, Watanabe T, Hatta M, Shinya K, Suresh M, Kawaoka Y, Rosen H, and Oldstone MB 2011. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proc Natl Acad Sci U S A 108: 12018–12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, van den Akker JPC, Molenkamp R, Koopmans MPG, van Gorp ECM, Haagmans BL, de Swart RL, Sette A, and de Vries RD 2020. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, Hippenstiel S, Dingeldey M, Kruse B, Fauchere F, Baysal E, Mangold M, Henze L, Lauster R, Mall MA, Beyer K, Rohmel J, Voigt S, Schmitz J, Miltenyi S, Demuth I, Muller MA, Hocke A, Witzenrath M, Suttorp N, Kern F, Reimer U, Wenschuh H, Drosten C, Corman VM, Giesecke-Thiel C, Sander LE, and Thiel A. 2020. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. [DOI] [PubMed] [Google Scholar]
- 45.Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, and Xing Z. 2020. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol 20: 615–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, and Clausen H. 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 32: 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, and Sette A. 2006. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC bioinformatics 7: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh S, Terabe M, Pendleton CD, Bhattacharyya A, Bera TK, Epel M, Reiter Y, Phillips J, Linehan WM, Kasten-Sportes C, Pastan I, and Berzofsky JA 2004. Human CTLs to wild-type and enhanced epitopes of a novel prostate and breast tumor-associated protein, TARP, lyse human breast cancer cells. Cancer Res 64: 2610–2618. [DOI] [PubMed] [Google Scholar]
- 49.Stuber G, Leder GH, Storkus WT, Lotze MT, Modrow S, Szekely L, Wolf H, Klein E, Karre K, and Klein G. 1994. Identification of wild-type and mutant p53 peptides binding to HLA-A2 assessed by a peptide loading-deficient cell line assay and a novel major histocompatibility complex class I peptide binding assay. Eur J Immunol 24: 765–768. [DOI] [PubMed] [Google Scholar]
- 50.Khan AA, Srivastava R, Spencer D, Garg S, Fremgen D, Vahed H, Lopes PP, Pham TT, Hewett C, Kuang J, Ong N, Huang L, Scarfone VM, Nesburn AB, Wechsler SL, and BenMohamed L. 2015. Phenotypic and functional characterization of herpes simplex virus glycoprotein B epitope-specific effector and memory CD8+ T cells from symptomatic and asymptomatic individuals with ocular herpes. J Virol 89: 3776–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srivastava R, Dervillez X, Khan AA, Chentoufi AA, Chilukuri S, Shukr N, Fazli Y, Ong NN, Afifi RE, Osorio N, Geertsema R, Nesburn AB, Wechsler SL, and BenMohamed L. 2016. The Herpes Simplex Virus Latency-Associated Transcript Gene Is Associated with a Broader Repertoire of Virus-Specific Exhausted CD8+ T Cells Retained within the Trigeminal Ganglia of Latently Infected HLA Transgenic Rabbits. J Virol 90: 3913–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan AA, Srivastava R, Vahed H, Roy S, Walia SS, Kim GJ, Fouladi MA, Yamada T, Ly VT, Lam C, Lou A, Nguyen V, Boldbaatar U, Geertsema R, Fraser NW, and BenMohamed L. 2018. Human Asymptomatic Epitope Peptide/CXCL10-Based Prime/Pull Vaccine Induces Herpes Simplex Virus-Specific Gamma Interferon-Positive CD107(+) CD8(+) T Cells That Infiltrate the Corneas and Trigeminal Ganglia of Humanized HLA Transgenic Rabbits and Protect against Ocular Herpes Challenge. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield C-GG, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, and Montefiori DC 2020. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X, Giorgi EE, Marichann MH, Foley B, Xiao C, Kong XP, Chen Y, Korber B, and Gao F. 2020. Emergence of SARS-CoV-2 through Recombination and Strong Purifying Selection. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng M, and Song L. 2020. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell Mol Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, and Veesler D. 2020. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tilocca B, Soggiu A, Musella V, Britti D, Sanguinetti M, Urbani A, and Roncada P. 2020. Molecular basis of COVID-19 relationships in different species: a one health perspective. Microbes Infect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, and Sette A. 2020. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 27: 671–680 e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee SS, and Chakraborty C. 2020. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J Med Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baruah V, and Bose S. 2020. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol 92: 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahmed SF, Quadeer AA, and McKay MR 2020. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, and Peters B. 2019. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47: D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sette A, and Sidney J. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50: 201–212. [DOI] [PubMed] [Google Scholar]
- 64.Sette A, and Sidney J. 1998. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr Opin Immunol 10: 478–482. [DOI] [PubMed] [Google Scholar]
- 65.Hertz T, and Yanover C. 2007. Identifying HLA supertypes by learning distance functions. Bioinformatics 23: e148–155. [DOI] [PubMed] [Google Scholar]
- 66.Chentoufi AA, Zhang X, Lamberth K, Dasgupta G, Bettahi I, Nguyen A, Wu M, Zhu X, Mohebbi A, Buus S, Wechsler SL, Nesburn AB, and BenMohamed L. 2008. HLA-A*0201-restricted CD8+ cytotoxic T lymphocyte epitopes identified from herpes simplex virus glycoprotein D. J Immunol 180: 426–437. [DOI] [PubMed] [Google Scholar]
- 67.Jespersen MC, Peters B, Nielsen M, and Marcatili P. 2017. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45: W24–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monto AS, DeJonge PM, Callear AP, Bazzi LA, Capriola SB, Malosh RE, Martin ET, and Petrie JG 2020. Coronavirus Occurrence and Transmission Over 8 Years in the HIVE Cohort of Households in Michigan. The Journal of infectious diseases 222: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, da Silva Antunes R, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, and Weiskopf D. 2020. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 370: 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, and Sette A. 2020. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schafer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo YM, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, and Crowe JE Jr. 2020. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584: 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ng OW, and Tan YJ 2017. Understanding bat SARS-like coronaviruses for the preparation of future coronavirus outbreaks - Implications for coronavirus vaccine development. Hum Vaccin Immunother 13: 186–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, and Jiang T. 2020. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 27: 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watanabe Y, Allen JD, Wrapp D, McLellan JS, and Crispin M. 2020. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369: 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin JE, Louder MK, Holman LA, Gordon IJ, Enama ME, Larkin BD, Andrews CA, Vogel L, Koup RA, Roederer M, Bailer RT, Gomez PL, Nason M, Mascola JR, Nabel GJ, Graham BS, and Team VRCS 2008. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine 26: 6338–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He Y, and Jiang S. 2005. Vaccine design for severe acute respiratory syndrome coronavirus. Viral Immunol 18: 327–332. [DOI] [PubMed] [Google Scholar]
- 77.He Y, Zhou Y, Siddiqui P, and Jiang S. 2004. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem Biophys Res Commun 325: 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sidney J, Peters B, Frahm N, Brander C, and Sette A. 2008. HLA class I supertypes: a revised and updated classification. BMC Immunol 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Correale P, Mutti L, Pentimalli F, Baglio G, Saladino RE, Sileri P, and Giordano A. 2020. HLA-B*44 and C*01 Prevalence Correlates with Covid19 Spreading across Italy. Int J Mol Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Correale P, Saladino RE, Giannarelli D, Sergi A, Mazzei MA, Bianco G, Giannicola R, Iuliano E, Forte IM, Calandruccio ND, Falzea AC, Strangio A, Nardone V, Pastina P, Tini P, Luce A, Caraglia M, Caracciolo D, Mutti L, Tassone P, Pirtoli L, Giordano A, and Tagliaferri P. 2020. HLA Expression Correlates to the Risk of Immune Checkpoint Inhibitor-Induced Pneumonitis. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crooke SN, Ovsyannikova IG, Kennedy RB, and Poland GA 2020. Immunoinformatic identification of B cell and T cell epitopes in the SARS-CoV-2 proteome. Scientific reports 10: 14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang LF, Anderson DE, Mackenzie JS, and Merson MH 2020. From Hendra to Wuhan: what has been learned in responding to emerging zoonotic viruses. Lancet 395: e33–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Plowright RK, Peel AJ, Streicker DG, Gilbert AT, McCallum H, Wood J, Baker ML, and Restif O. 2016. Transmission or Within-Host Dynamics Driving Pulses of Zoonotic Viruses in Reservoir-Host Populations. PLoS Negl Trop Dis 10: e0004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eng CL, Tong JC, and Tan TW 2016. Distinct Host Tropism Protein Signatures to Identify Possible Zoonotic Influenza A Viruses. PloS one 11: e0150173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simons RR, Gale P, Horigan V, Snary EL, and Breed AC 2014. Potential for introduction of bat-borne zoonotic viruses into the EU: a review. Viruses 6: 2084–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, da Silva Antunes R, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, and Weiskopf D. 2020. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, and Sette A. 2020. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181: 1489–1501 e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baggio S, L’Huillier AG, Yerly S, Bellon M, Wagner N, Rohr M, Huttner A, Blanchard-Rohner G, Loevy N, Kaiser L, Jacquerioz F, and Eckerle I. 2020. SARS-CoV-2 viral load in the upper respiratory tract of children and adults with early acute COVID-19. Clin Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moratto D, Giacomelli M, Chiarini M, Savare L, Saccani B, Motta M, Timpano S, Poli P, Paghera S, Imberti L, Cannizzo S, Quiros-Roldan E, Marchetti G, and Badolato R. 2020. Immune response in children with COVID-19 is characterized by lower levels of T-cell activation than infected adults. Eur J Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, Lubke M, Bauer J, Rieth J, Wacker M, Peter A, Horber S, Traenkle B, Kaiser PD, Rothbauer U, Becker M, Junker D, Krause G, Strengert M, Schneiderhan-Marra N, Templin MF, Joos TO, Kowalewski DJ, Stos-Zweifel V, Fehr M, Rabsteyn A, Mirakaj V, Karbach J, Jager E, Graf M, Gruber LC, Rachfalski D, Preuss B, Hagelstein I, Marklin M, Bakchoul T, Gouttefangeas C, Kohlbacher O, Klein R, Stevanovic S, Rammensee HG, and Walz JS 2020. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. [DOI] [PubMed] [Google Scholar]
- 91.Ong EZ, Chan YFZ, Leong WY, Lee NMY, Kalimuddin S, Haja Mohideen SM, Chan KS, Tan AT, Bertoletti A, Ooi EE, and Low JGH 2020. A Dynamic Immune Response Shapes COVID-19 Progression. Cell Host Microbe 27: 879–882 e872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, Chia WN, Chen MI, Wang LF, Ooi EE, Kalimuddin S, Tambyah PA, Low JG, Tan YJ, and Bertoletti A. 2020. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584: 457–462. [DOI] [PubMed] [Google Scholar]
- 93.Welsh RM, and Selin LK 2002. No one is naive: the significance of heterologous T-cell immunity. Nat Rev Immunol 2: 417–426. [DOI] [PubMed] [Google Scholar]
- 94.Becker MM, Graham RL, Donaldson EF, Rockx B, Sims AC, Sheahan T, Pickles RJ, Corti D, Johnston RE, Baric RS, and Denison MR 2008. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci U S A 105: 19944–19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The human specific SARS-CoV-2 complete genome sequences were retrieved from the GISAID database, whereas the SARS-CoV-2 sequences for pangolin (Manis javanica), and bat (Rhinolophus affinis, Rhinolophus malayanus) were retrieved from NCBI. Genome sequences of previous strains of SARS-CoV for human, bat, civet, and camel were retrieved from the NCBI GenBank.