Fig. 3. Overall features of the RNA around the most preferred APOBEC-edited sites on SARS-CoV-2.
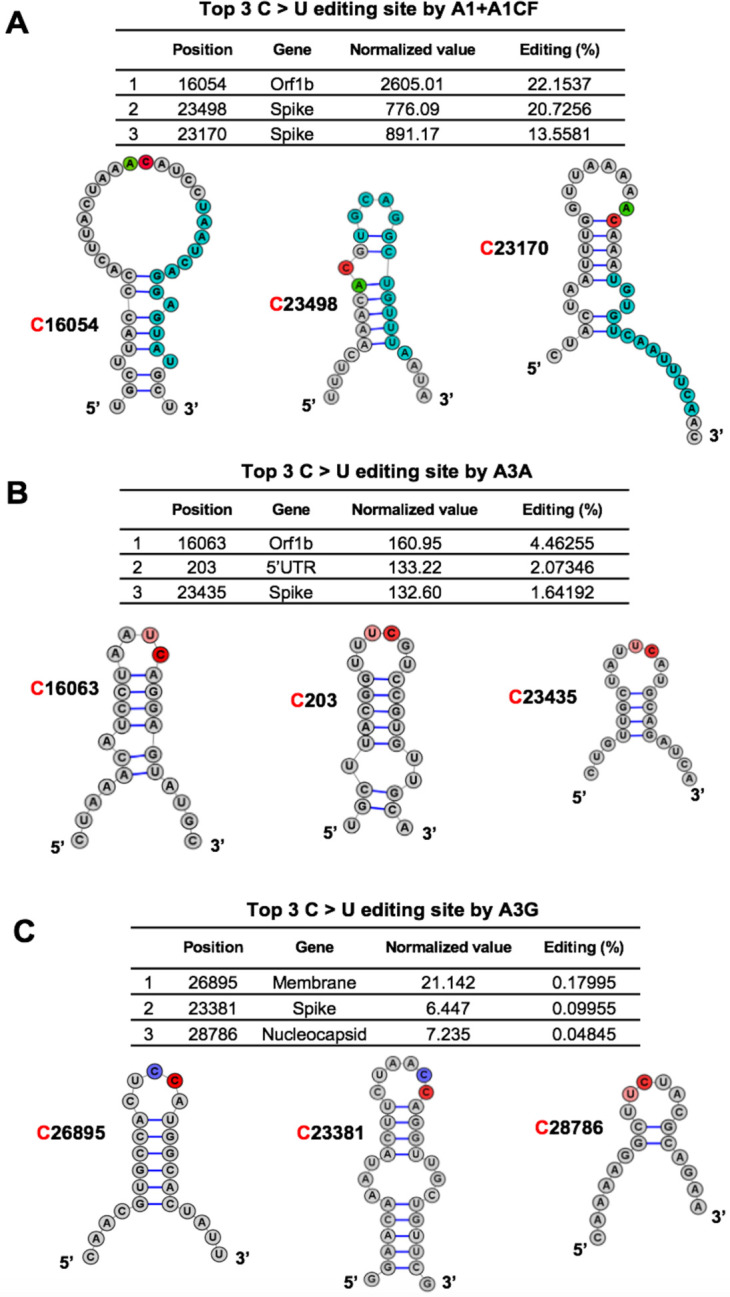
The predicted RNA secondary structures (55) of the sequences near the top 3 highest editing C sites by A1+A1CF (A), A3A (B), and A3G (C) (See related Table S2). The editing efficiency of each site is listed at the top of each panel. In the secondary structure, the target C sites are highlighted in red, and −1 positions of the target C sites are highlighted in green for A, pink for U, and blue for C, respectively. In panel-A, the proposed canonical mooring sequences for A1+A1CF (highlighted in sky blue) contain relatively high U/A/G contents downstream of the target C.
