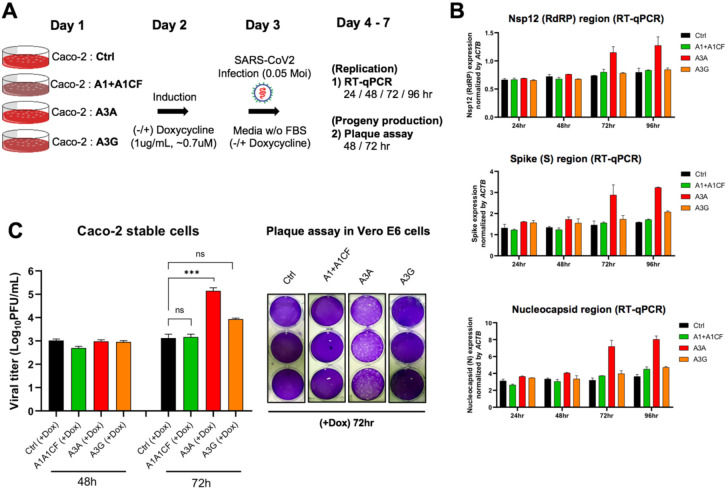
Fig. 5. SARS-CoV-2 replication and virion production in cells expressing APOBECs.
(A) Overview of experiments for SARS-CoV-2 replication and viral production in the presence of APOBECs. The Caco-2 stable cell lines were constructed to express A1+A1CF, A3A, or A3G under a tetracycline-controlled promoter. The Caco-2-APOBEC stable cell lines were then infected with SARS-CoV-2 (MOI = 0.05), and the viral RNA replication and progeny production were measured at different time points. (B) Effect of each APOBEC expression on SARS-CoV-2 viral RNA replication. Measurement of relative viral RNA abundance at different time points after viral infection of the Caco-2-APOBEC stable cell lines expressing A1+A1CF, A3A, or A3G. The viral RNA abundance was measured using real-time quantitative PCR (qPCR) to detect RNA levels by using specific primers to amplify three separate viral regions, the Nsp12, S, or N coding regions (see Methods in SI). (C) Effect of each APOBEC expression on SARS-CoV-2 progeny production. Infectious viral progeny yield harvested in the medium at 48 hrs and 72 hrs post-infection was determined by plaque assay (see Methods). Statistical significance was calculated by unpaired two-tailed student’s t-test with P-values represented as: P > 0.05 = not significant, *** = P < 0.001.