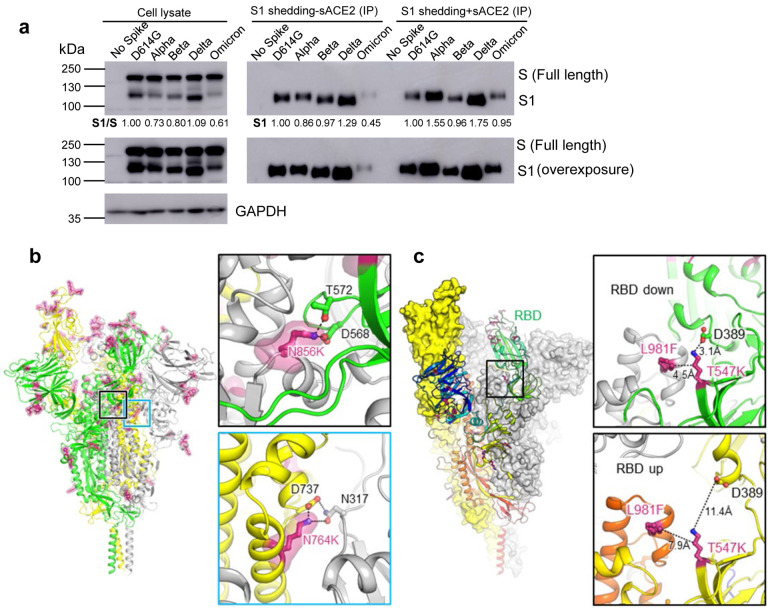
Fig. 3. The Omicron S protein exhibits low S1 shedding, consistent with predicted increase in stability.
(a) HEK293T cells were transfected with S constructs and treated with or without sACE2. Cell lysate and cell culture media were collected after sACE2 treatment, and shed S1 subunit in cell culture media was immune-precipitated with anti-Flag beads. Blots were probed with anti-S1 and anti-GAPDH, and S1 shedding was quantified by NIH ImageJ by setting D614G to 1.0. (b) Structure of Omicron spike protein viewed from side. Mutations of Omicron are highlighted by red sticks and semi-transparent red surfaces. RBD of the yellow protomer is in an “up” conformation. Upper inset: the mutation N856K enables formation of salt-bridge and hydrogen bond with the residues D568 and T572, respectively on the neighboring protomer (green). Lower inset: the mutation N764K enables formation of hydrogen bond with residue N317 on the neighboring protomer (grey) and salt-bridge with residue D737 on the same protomer (yellow). (c) Omicron mutations T547K and L981F stabilize RBD in “down” conformation. Structure of Omicron spike protein by homology modeling was illustrated as surfaces (yellow and grey protomer) and ribbon (rainbow protomer). Upper inset: When RBD is in “down” conformation, Omicron mutation T547K enables formation of salt-bridge with residue D389 located on the base of RBD. Mutation T547K and L981F together enhance the hydrophobic interaction between the neighboring protomers (green and grey). Lower inset: When RBD is in “up” conformation, the interactions between these residues are disrupted.