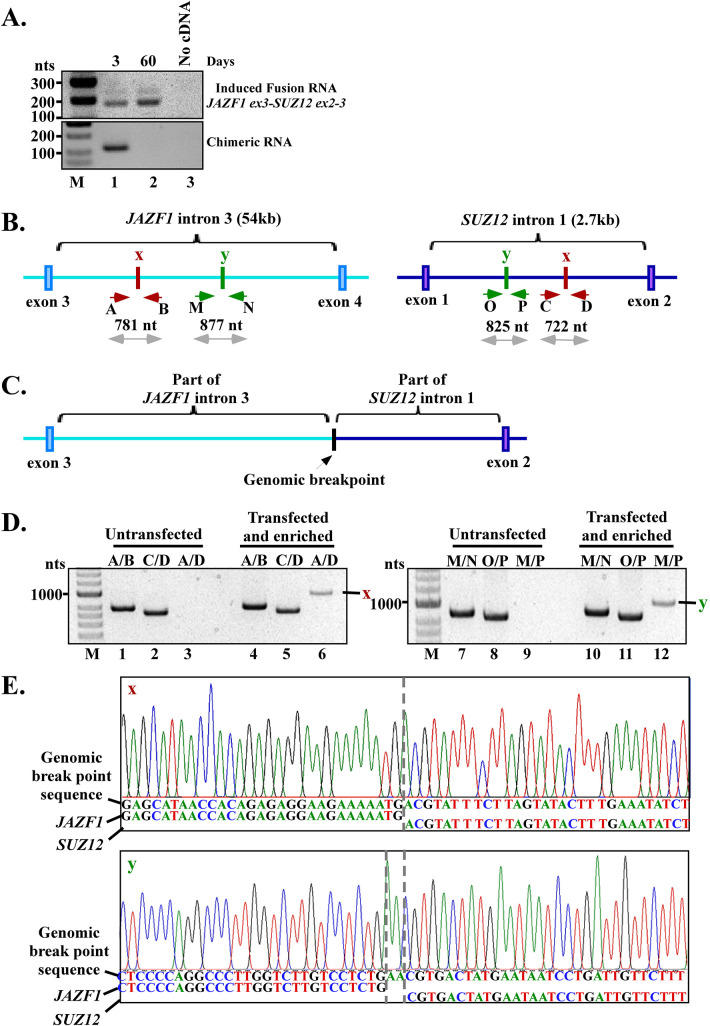
Fig 4. Induced JAZF1-SUZ12 fusion is the result of genomic rearrangements.
(A) RT-PCR shows the transient nature of exogenously expressed chimeric RNA that was degraded and completely absent by day 60 (lower panel, lane 1 vs. 2), and the persistent nature of the induced fusion transcript (upper panel, lane 1 vs. 2) which was continuously expressed up to day 60 in the enriched cell population. This shows that the continuously expressed JAZF1-SUZ12 fusion RNA does not require the presence of chimeric RNAs. See S5 Fig for procedures used to propagate and enrich the induced hESC population. (B) The wild-type alleles with two identified genomic breakpoints marked as ‘x’ and ‘y’, and the primers used to amplify these breakpoints. The intron sizes are not presented in proportion as JAZF1 intron-3 (54kb) is much larger than SUZ12 intron-1 (2.7kb). (C) Schematics of the rearranged allele of the final fusion gene with JAZF1 and SUZ12 joined at the intron breakpoints. (D) The unrearranged wild-type JAZF1 allele near breakpoint ‘x’ was amplified by primer pair A/B (781 bp; lanes 1 and 4) and near breakpoint ‘y’ by primer M/N (877 bp; lanes 7 and 10). The unrearranged wild-type SUZ12 allele near breakpoint ‘x’ was amplified by primer pair C/D (722 bp; lanes 2 and 5) and near breakpoint ‘y’ by primer O/P (825 bp; lanes 8, and 11). The genomic fusion band ‘x’ (951 bp) revealed by fusion-specific primer pair A/D, and fusion band ‘y’ (976 bp) revealed by primer pair M/P, were present only in the enriched hESC population but absent in untransfected hESC cells (lane 6 vs. 3, and lane 12 vs. 9). See S6 Fig for multiplex primer designs used for initial scanning of potential breakpoints. (E) Sanger sequencing of the ‘x’ and ‘y’ fusion band identified the exact genomic breakpoints marked by dash lines. The genomic breakpoint ‘y’ contains a ‘AA’ insertion (marked by dash lines). The full-length Sanger sequences of 951 bp for “x” and 976 bp for “y” are shown in S7 and S8 Figs.