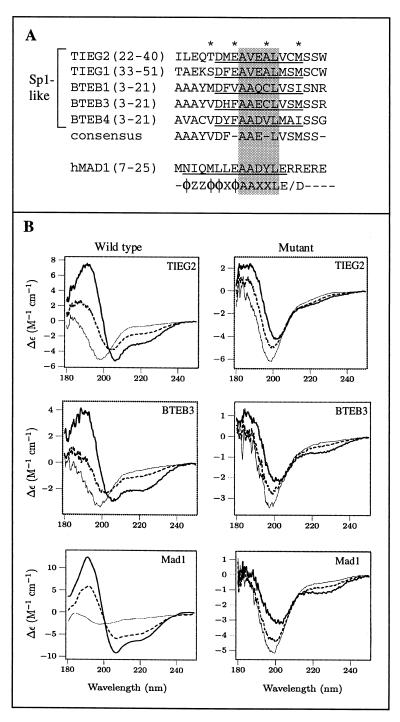
FIG. 6.
A conserved α-helical motif is present in TIEG and BTEB proteins. (A) Sequence alignment of regions in the TIEG and BTEB proteins homologous to TIEG2 R1. Identical residues within this region of these Sp1-like proteins are indicated by asterisks. The amino acid residues corresponding to the conserved α-helical region are underlined. The consensus sequence is shown below. For comparison, the mouse Mad1 SID, along with the PAH2 interaction motif, as defined by Brubaker et al. (4), is included. Note that the sequences outside the AAXXL core (shaded residues) differ significantly between the Sp1-like proteins and the Mad1 SID. (B) CD analysis of peptides containing consensus sequences from TIEG2 R1, BTEB3, the Mad1 SID, and their respective mutant forms was performed as described in Materials and Methods. Note that the wild-type peptides are random coils in the absence of TFE (thin line) but become helical in the presence of 20% (dashed line) and 50% (thick line) TFE, as evidenced by the strong negative peaks at 208 and 222 nm. This trend is not observed in the mutant peptides, which remain largely unstructured under the same conditions. The spectrum of each peptide represents the average of three scans. Smoothing was performed for the wild-type Mad1 SID only.