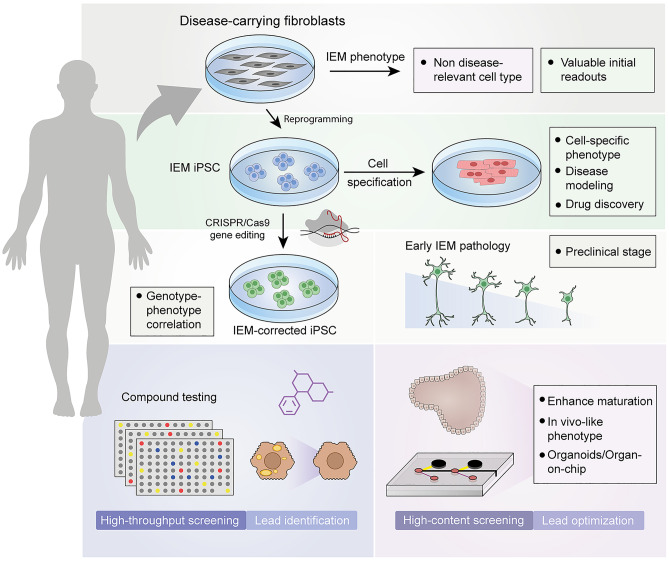
Fig. 4.
Schematic representation of the lessons learnt from iPSC models of IEMs. Somatic cells such as fibroblasts can be obtained from patients with IEMs to study pathogenic mechanisms, but in a cellular and metabolic context that may differ from that of disease-affected cells. Somatic cells can be reprogrammed into iPSC carrying the IEM-specific genetic alteration. iPSC can be differentiated into the disease-relevant cell type (such as cardiomyocytes to study disorders of energy metabolism or glycogen storage disorders). CRISPR/Cas9-based gene-editing technology allows the generation of isogenic sets of iPSC that can be used to properly associate genetic alterations to specific phenotypes. The combination of gene editing and iPSC technology is a remarkable tool to study preclinical stages (such as the neural defects in Sanfilippo C) and to perform high-throughput drug screenings (for example to reduce the hypercholesterolemia in iPSC-derived hepatocytes). The immature phenotype of iPSC derivatives is a current limitation in the field, which can be overcome by using advanced culture systems such as organoids and organ-on-a-chip devices, which generate cells more similar to their in vivo counterpart, making them suitable for high-content screenings and lead optimization