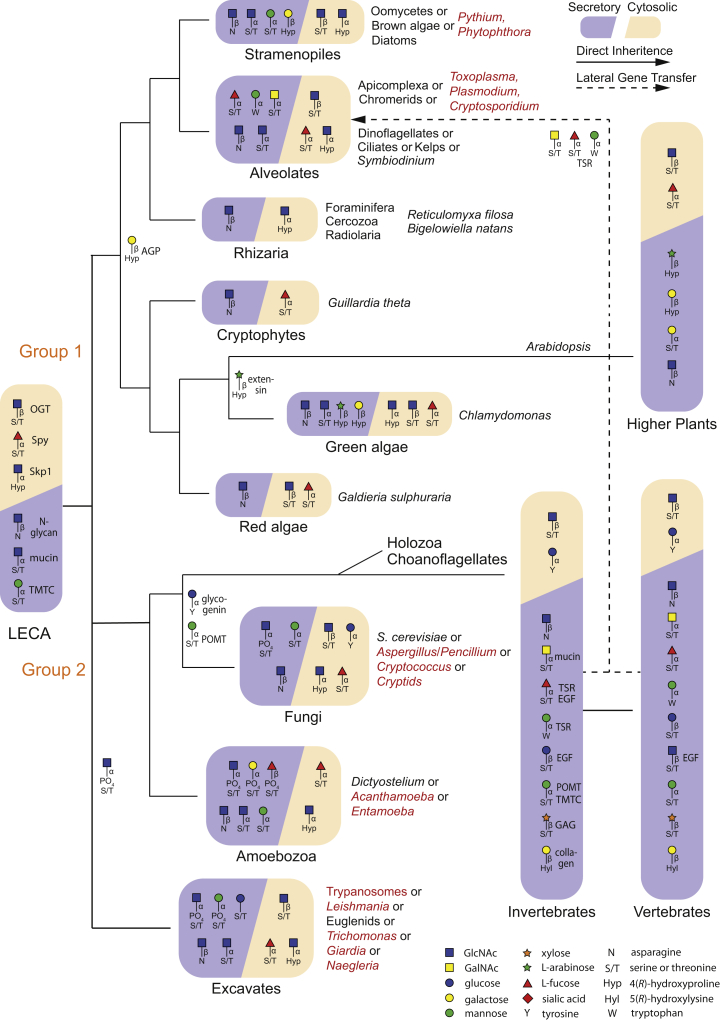
Fig. 1.
Evolutionary map of eukaryotic glycans based on the core sugar–amino acid linkage type. Glycan protein linkages, based on direct evidence or inferred from glycogenomics (see text), are layered on a cladogram from a current model for the phylogeny of eukaryotes (3), which emphasizes the protist subkingdoms and their proposed relation to the last eukaryotic common ancestor (LECA). Major groups where glycomic or bioinformatics information is available are named, with species names in italics. Human or plant pathogens are in red. Linkages found in any one species qualifies for assignment in the group. The dashed line represents likely lateral gene transfer. Poorly studied protists for which there is a lack of experimental data are not shown. Organisms that branched from the lineage that gave rise to the higher plants are referred to as Group 1, whereas those that gave rise to animals are classified as Group 2. Linkages are inferred to occur in the LECA if they are found in both groups of protists, but the absence of a linkage might be a result of incomplete information. The origin of linkages inferred to originate after the LECA is shown at the relevant branch point. Notes adjacent to linkages indicate names by which they are commonly referred. GPI anchors, a specialized glycolipid linked to protein C termini typically via a phosphoethanolamine linker to a nonreducing terminal mannose, was likely present in the LECA (not shown). SNFG symbols for sugars are summarized at the bottom. See text for explanations.