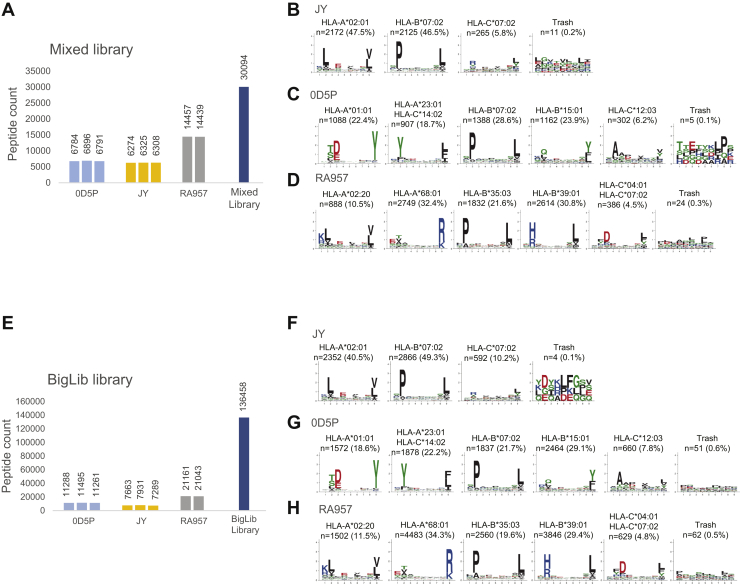
Fig. 3.
Application of mixed and BigLib multi-HLA spectral libraries for matching immunopeptidomics DIA data. DIA data of JY, OD5P, and RA957 samples were matched against the mixed library comprising immunopeptidomics data from six different biological samples (A). The number of peptides identified in each of the DIA measurements and the number of peptides included in the mixed library are reported. Deconvolution of the consensus binding motifs of JY (B), OD5P (C), and RA957 (D) DIA immunopeptidomics samples with MixMHCp 2.1 and manual annotation of motifs. The number of peptides and the HLA restriction assigned to each motif are reported. DIA data of JY, OD5P, and RA957 samples were matched against the BigLib library comprising immunopeptidomics data from 40 different biological samples (E). The number of peptides identified in each of the DIA measurements and the number of peptides included in the mixed library are reported. Deconvolution of the consensus binding motifs of JY (F), OD5P (G), and RA957 (H) DIA immunopeptidomics samples as described above. DIA, data-independent acquisition; HLA, human leukocyte antigen.