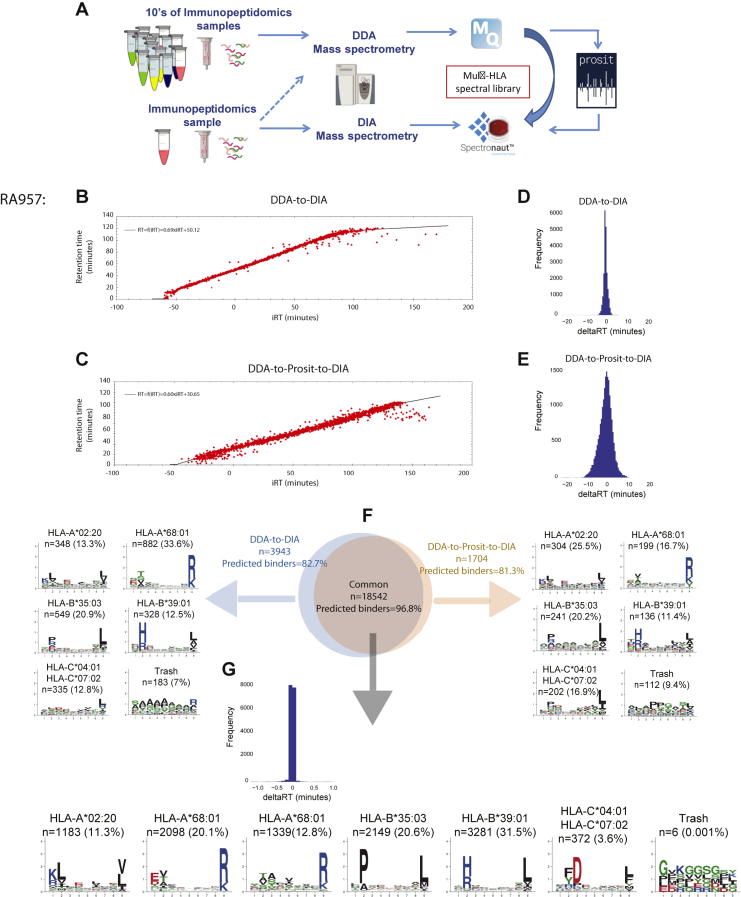
Fig. 5.
Overview of the performance of Prosit-predicted MS/MS spectral library. A schematic overview of our approach to estimate the performance of Prosit-predicted MS/MS spectral library of all HLA peptides included in the BigLib that were used to match DIA data of RA957 samples (A). Correlations between measured RT and the predicted RT calculated by Spectronaut (RT = f(iRT)) of peptides identified in RA957 applying the BigLib library in the DDA-to-DIA approach (B) and with the Prosit-predicted BigLib library in the DDA-to-Prosit-to-DIA approach (C). The difference between the predicted (RT =f(iRT)) and the mean measured RT in the DIA data calculated for peptides identified in the DDA-to-DIA (D) and in the DDA-to-Prosit-to-DIA (E). The overlap between peptides identified in the two approaches is represented in a Venn diagram (F). The fraction of peptides predicted as HLA binders are provided for unique and common peptide groups. Nine mer peptides from each group were clustered to reveal the binding motifs. Frequency plot of the delta apex, which is the difference between the measured peptide apex obtained in the DDA-to-DIA and the DDA-to-Prosit-to-DIA (G). DDA, data-dependent MS/MS acquisition; DIA, data-independent acquisition; HLA, human leukocyte antigen; MS/MS, tandem MS; RT, retention time.