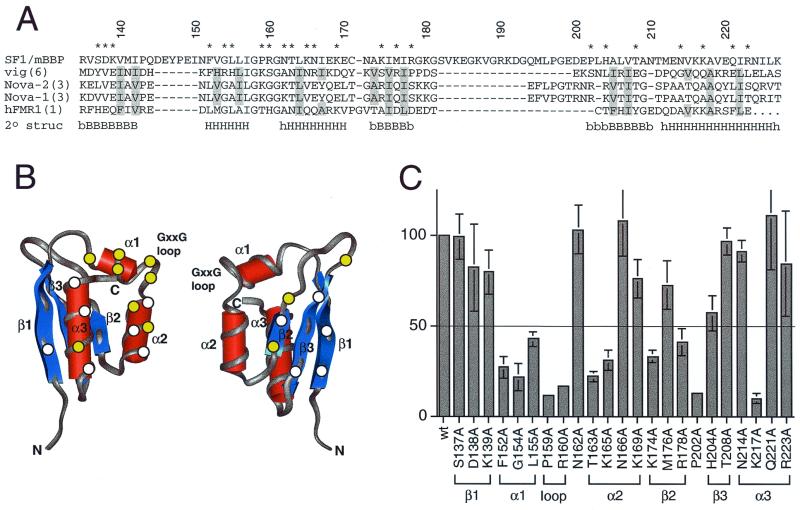
FIG. 5.
Activation of the BPS reporter by Tat-fused SF1/mBBP mutants. (A) Sequence alignment of the maxi-KH domain of SF1/mBBP and selected KH domains from vigilin (vig; domain 6), Nova-2 (domain 3), Nova-1 (domain 3), and hFMR-1 (domain 1). Secondary structure elements were assigned based on the X-ray and NMR structures of these individual domains (34, 39; Musco et al., letter). B and H, β-sheet and α-helical residues, respectively, which are defined clearly in all structures; lowercase letters, residues that can be assigned to a secondary structure in only one or two structures. Residues considered to be part of the hydrophobic core and therefore not chosen for mutation are shaded. Numbers refer to the SF1/mBBP sequence, and residues marked with an asterisk were mutated to alanines. (B) Averaged NMR structure of the sixth KH domain from vigilin (PDB file 1VIH [39]), shown from two different faces. Circles, approximate locations of amino acids chosen for mutagenesis, assuming that SF1/mBBP adopts a similar fold; yellow circles, positions that decrease activity by at least twofold (see panel C); white circles, positions that have little or no effect. (C) Activities of the Tat-fused SF1/mBBP mutants normalized to the activity of the wild-type (wt) protein with standard deviations (bars) calculated as for Fig. 3. The line corresponds to a twofold decrease in activity. The predicted corresponding units of secondary structures are indicated.