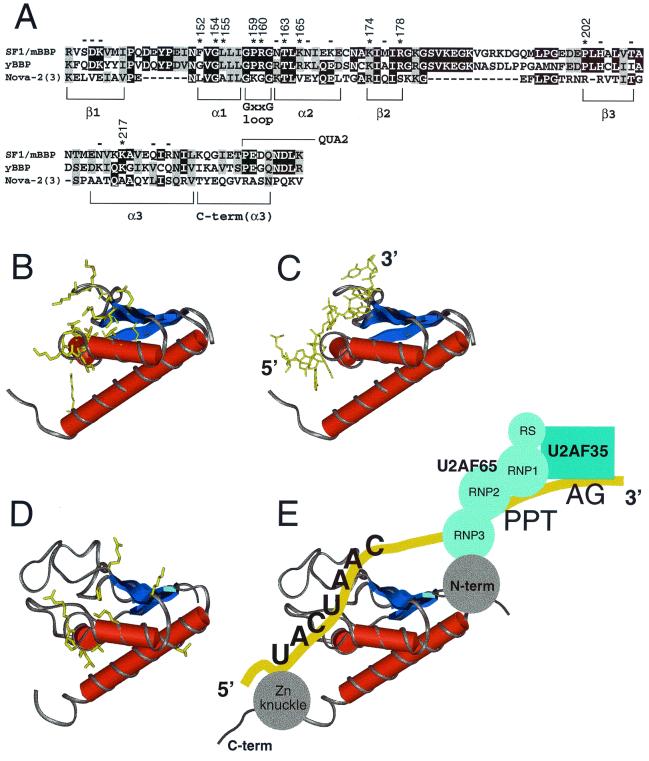
FIG. 6.
(A) Sequence alignment of SF1/mBBP, BBP from Saccharomyces cerevisiae (yBBP), and KH domain 3 from Nova-2. Black boxes, identical residues; shaded boxes, conserved residues. The corresponding units of secondary structure are indicated. An extra C-terminal region is shown, compared to the sequence shown in Fig. 5A; this region corresponds to part of the Nova-2 structure. Asterisks and numbers, positions in SF1/mBBP important for binding based on the mutagenesis data (Fig. 5); minuses, positions where mutations have little or no effect. The numbering of Nova-2 residues discussed in the text (and not shown) corresponds to that described for the cocrystal structure (35). The region corresponding to the beginning of QUA2 is indicated. (B) Structure of the Nova-2 KH domain, taken from the cocrystal structure of the protein-RNA complex (35). Amino acids involved in RNA binding are shown in yellow. (C) Structure of the Nova-2 KH domain complexed to RNA (35), with the RNA tetranucleotide specifically recognized by the protein shown in yellow. The 5′ and 3′ ends of the RNA are indicated. (D) Homology model of SF1/mBBP maxi-KH domain, based on the structure of the RNA-bound Nova-2 domain and the alignment shown in panel A. The approximate locations of residues important for RNA binding, as defined by mutagenesis, are shown in yellow. The large β1/α1 and β2/β3 loops found in the SF1/mBBP maxi-KH domain were positioned arbitrarily by the homology modeling. (E) Proposed RNA-binding orientation of the SF1/mBBP-U2AF complex, based on similarities to the Nova-2 protein-RNA complex (see Discussion). The schematic drawing is not intended to indicate the relative orientations of the U2AF65 and U2AF35 subunits or of the RS domain of U2AF65, which also may contact the RNA (56).