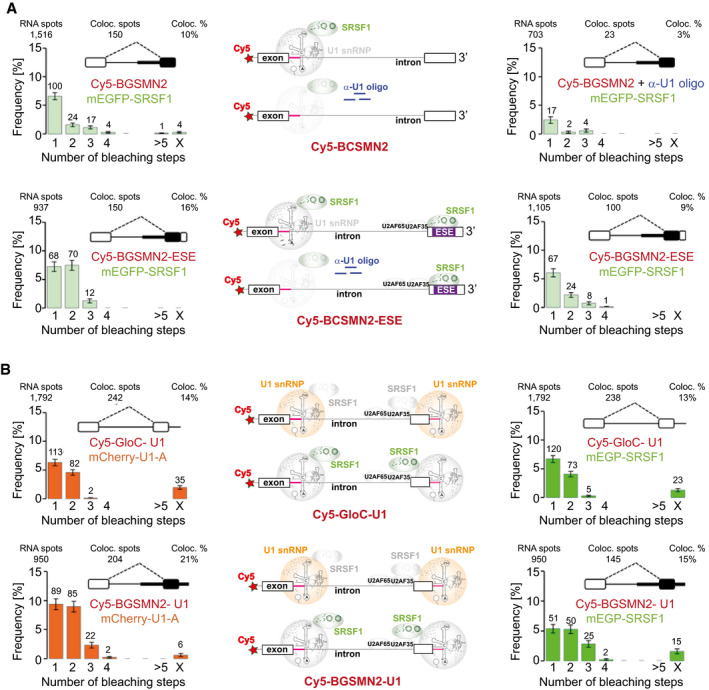
Effects of 3′‐terminal ESE repeats. Top left, two‐way colocalization of complexes formed in NE2 on BGSMN2, a chimeric globin/SMN2 pre‐mRNA. The heavy black line and box indicate the intron and exon sequences of SMN2 exon 7. Top right, the effects of inclusion of a 2′‐
O‐methyl oligonucleotide complementary to the 5′ end of U1 snRNA. The interpretative cartoons (top centre) show (upper) that most SRSF1/pre‐mRNA complexes contain a single molecule of SRSF1 and (lower) that the proportion of SRSF1‐associated pre‐mRNA molecules was reduced sharply by occlusion of the 5′ end of U1 snRNA by the oligonucleotides. Below, the corresponding experiments after addition of four tandem repeats of a strong ESE to the 3′ terminus (Jobbins
et al,
2018). Bottom left, most SRSF1/pre‐mRNA complexes contain two molecules of SRS1. In the interpretative cartoon in the centre (upper), this is shown by association of one SRSF1 with the U1 snRNP and the other with the ESEs (blue). We have shown previously that the activity of ESEs is limited by their occupancy, consistent with transient binding by SRSF1 and stabilization of one molecule by direct interactions based on diffusional encounters with splice site‐associated factors, since their effects are not blocked by the incorporation of intervening non‐RNA linkers (Jobbins
et al,
2018). Bottom right, double occupancy of the pre‐mRNA was greatly reduced by occlusion of the 5′ end of U1 snRNA, suggesting that the U1‐dependent but not ESE‐dependent association of SRSF1 was lost (lower cartoon). The error bars in the histograms are the square root of the variance of the binomial probability that an RNA spot will be associated with the given number of protein bleaching steps.