Figure 1.
Virus-dependent bivalent versus monovalent C10 neutralization differences
(A) The DENV2 sE dimer color-coded by domain with the C10 footprint outlined (heavy chain in magenta, light chain in cyan) (PDB: 4UT9). A green oval at the center marks the molecular 2-fold axis. The sE protomer contributing the fusion loop (FL; orange) or domain III (blue) to the epitope are dubbed “reference” or “opposite” subunit, respectively.
(B) The flavivirus mature virion displayed in surface representation with selected icosahedral symmetry axes shown as full green symbols: 2-fold (I2, oval), 3-fold (I3, triangle), and 5-fold (I5, pentagon). 90 E dimers are arranged as 30 “rafts” (green outlines) made of three E dimers. The central, I2 dimer (white) is flanked by two L2 dimers, (light/dark gray) formed about local 2-fold axes (open green ovals). The location of the C10 epitopes is outlined in magenta in the front raft, labeled 2f, 3f, and 5f.
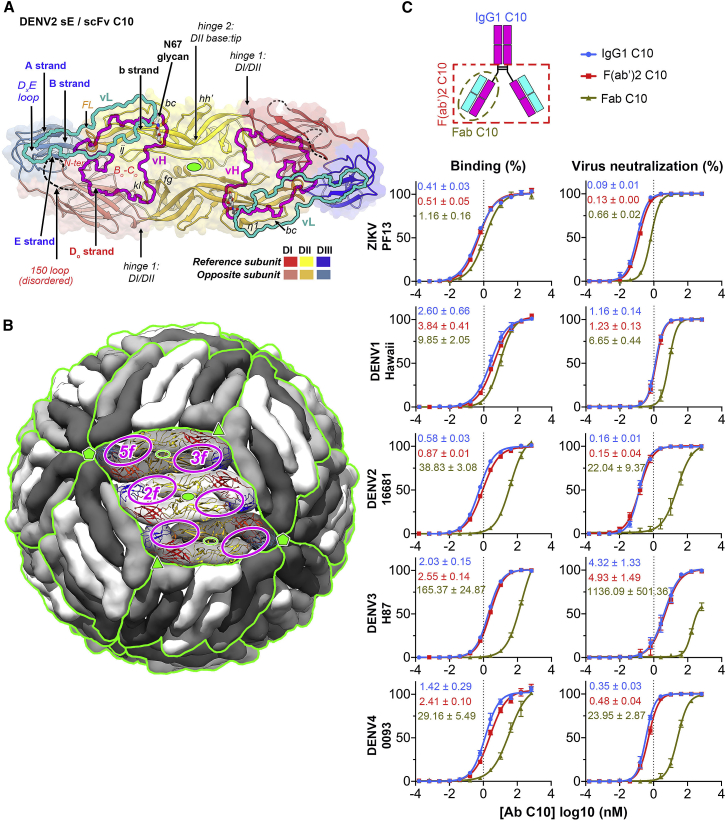
(C) Bivalent versus monovalent C10 binding and neutralization. Top: cartoon of IgG1, F(ab’)2, and Fab molecules. Bottom: ELISA titration curves and the estimated apparent dissociation constant (KD) (left panels) and neutralization curves with the 50% focus reduction neutralization titer (FRNT 50%) measured on Vero cells (right panels) for ZIKV and DENV1–DENV4 grown in insect cells. Data are from three independent experiments.