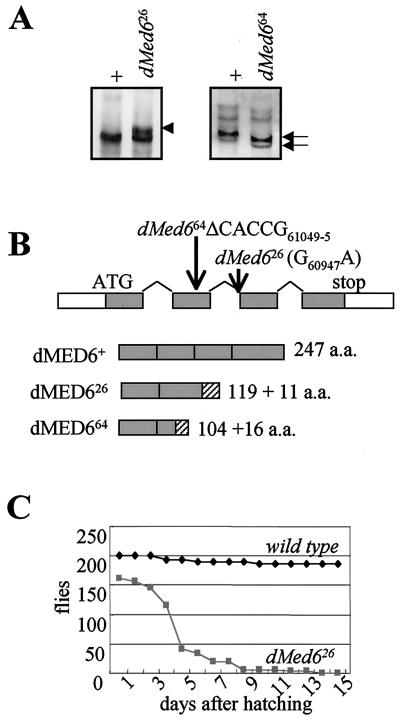
FIG. 3.
Identification of dMed6 mutations. (A) SSCP analysis of dMed6 mutant alleles. PCR fragments (positions 61159 to 60748) amplified from wild-type (+), dMed626, and dMed664 mutant chromosomes were analyzed by SSCP gel electrophoresis. The extra band (arrowhead) and faster-migrating bands (arrows) are marked. (B) Mutation sites of dMed626 and dMed664. The structure of the dMed6 gene is marked with the mutations in each dMed6 mutant allele identified. dMed626 has one nucleotide change (G60947A) at the splicing acceptor of the second intron, and dMed664 has a five-nucleotide deletion (ΔCACCG61049–5). Both mutations cause truncation of the dMED6 protein, as shown beneath. Gray boxes, wild-type coding regions; hatched boxes, nonauthentic amino acids added to the C-terminal ends of the mutant proteins due to the mutations. (C) Lethality of dMed6 homozygous mutants. The numbers of larvae, pupae, and flies viable after hatching from eggs were determined every day for wild-type (200 hatched larvae) and dMed626 mutant flies (168 hatched larvae) at 25°C. Most of the wild-type flies developed to adults in 10 days after hatching. However, a significant number of mutant flies died 3 to 5 days after hatching. The mutant flies showed no apparent developmental defect until the third larval instar but never developed to the prepupa stage.