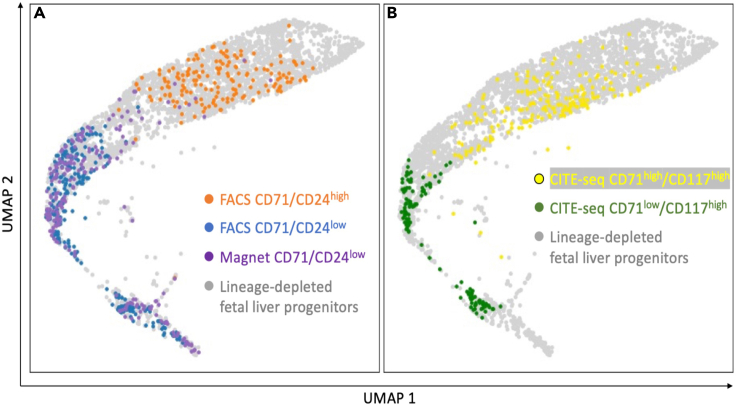
Figure 4.
Single-cell RNA-seq analysis of erythroid progenitor populations
(A) Single cell RNA sequencing was performed using the 10× Genomics 3′ Digital Gene Expression platform on lineage depleted fetal liver progenitor cells and (A) FACS-isolated CD71/CD24low and CD71/CD24high cells and magnetically isolated CD71/CD24low cells.
(B) For comparative reference, proteogenomic oligo-tagged CITE-seq (Stoeckius et al., 2017) analysis of CD71 and CD117 expression in lineage depleted fetal liver progenitors identifies transcriptome state of individual CD71low/CD117high and CD71high/CD117high erythroid progenitor cells. TotalSeqA0012 anti-mouse CD117 and TotalSeqA0441 anti-mouse CD71 antibodies (Biolegend) were used for CITE-seq staining. scRNAseq output was analyzed and visualized by the dimensionality reduction approach of Universal Manifold Approximation and Projection (UMAP) (Becht et al., 2018).