Highlights
-
•
A newly developed inhibitor, PAKib suppresses the kinase activity of PAK4 specifically with IC50 around 500 nM.
-
•
PAKib inhibits pancreatic cancer cell growth in vitro and in a syngeneic mouse model of pancreatic cancer.
-
•
PAKib stimulates the inhibitory effect of gemcitabine on pancreatic cancer cell growth in vitro and in a syngeneic mouse model of pancreatic cancer.
-
•
PAKib suppresses pancreatic cancer cell growth via inducing apoptosis and cell death.
Keywords: Pancreatic cancer, PAK4, KRas, Gemcitabine
Abbreviations: PAK, p-21 activated kinase; PDA, pancreatic ductal adenocarcinoma
Abstract
Over 95% of Pancreatic ductal adenocarcinomas (PDA) carry mutations in the oncogene KRas which has been proven to be a difficult drug target. P21-activated kinase 4 (PAK4), acts downstream of KRas, and is overexpressed in PDA contributing to its growth and chemoresistance, and thus becomes an attractive therapeutic target. We have developed a new PAK4 inhibitor, PAKib and tested its effect on pancreatic cancer (PC) cell growth in vitro and in a syngeneic mouse model of PC. PAKib suppressed PC cell growth by inducing cell death and cycle arrest. PAKib inhibited PC growth and enhanced the inhibition by gemcitabine of PC in cell culture and in PC mouse model. PAKib acted through multiple signaling pathways involved in cell cycle checkpoints, apoptosis, cell junction, and focal adhesion. These proof-of-concept studies demonstrated the anti-cancer effect of PAKib alone and in combination with gemcitabine and warrant a further clinical investigation.
Introduction
Pancreatic ductal adenocarcinoma (PDA) remains one of the deadliest diseases, with a 5-year survival <10% for decades [1], largely due to lack of effective treatments. Over 95% of PDA carry mutations in the oncogene KRas [2], which is a difficult druggable target [3]. P21-activated kinases (PAKs) act downstream of KRas. PAK1 and PAK4, overexpressed and/or hyperactivated in PDA, contribute to the growth and metastasis of PDA [4], [5], [6], and to chemotherapeutic resistance of PDA [7], [8], [9]. Reduction in activity of both PAK1 and PAK4 increases sensitivity of PDA to gemcitabine chemotherapy in vitro and in vivo [10,11]. Inhibition of PAK1 and PAK4 stimulated anti-tumor immunity by increasing tumor infiltrating lymphocytes and decreasing PD-L1 (programed death-ligand 1) expression by pancreatic cancer (PC) cells [12]. These findings have highlighted the prominence of PAKs in PDA and indicated that targeting PAKs would present an exciting therapeutic opportunity for PC treatment.
Several PAK inhibitors have been shown to suppress pancreatic cancer in cell culture and in mouse models [4,8,9]. However, none have successfully gone through clinical trials due to off-target effects [6] or unknown causes. In pancreatic cancer, ATP-competitive inhibitors selective for PAK1 (FRAX597) and PAK4 (PF3758309) have been shown to suppress tumor growth in cell culture and mouse models [9,11]. However, the FRAX-related compounds also show strong adverse inhibition of hERG potassium channels, and they have poor cellular permeability, which blocked their passage through early clinical trials in other types of disease [13]. PF-3758309 failed in an early-phase clinical study in PDA due to unwarranted pharmacokinetic effects [6]. In addition, in pre-clinical settings, both FRAX597 and PF3758309 have been demonstrated to enhance the effects of chemotherapeutic agents currently used in clinic [9,11], indicating the application of PAK inhibition in combination with chemotherapy. Recently developed PAK4 allosteric modulators, KPT-9274 and its analogues, have been shown to inhibit PDA in cell-based and xenografted tumour mouse models, and KPT-9274 has been entered in an early-phase clinical trial (NCT02702492) [8]. The preclinical results from the development of PAK inhibitors are encouraging. Positioned downstream of difficult-to-target oncogenes such as Ras, PAKs remain promising druggable therapeutic targets. PAK inhibitors should open a new avenue for disruption of Ras-driven tumorigenesis as is the case with PDA.
We have developed a novel PAK4 inhibitor, PAKib. We have reported here that PAKib inhibited the activity of PAK4, that PAKib suppressed pancreatic cancer growth and further enhanced the inhibitory effect of gemcitabine on pancreatic cancer in vitro and in a mouse model, that PAKib acted through multiple signaling molecules involved in cell cycle checkpoints, apoptosis, cell junction, and focal adhesion.
Materials and methods
Reagents and cell culture
The human pancreatic cancer (PC) cell lines PANC-1, MiaPaCa-2 and BxPC3 were purchased from the American Type Culture Collection. The murine pancreatic cancer cell lines KPCPAK1WT833 was isolated and characterized from KPC mice as described previously [12]. PAKib was synthetized and provided by PAKinax Pty. Ltd (Melbourne, Australia). Gemcitabine was purchased from Sigma-Aldrich (Sydney, Australia). Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 5% fetal bovine serum (FBS) (Hyclone Laboratories, Melbourne, Australia) in a 37°C incubator with a humidified atmosphere of 5% CO2. p21 activated kinase 4 (PAK4) recombinant protein was purchased from MyBioSource, delivered by Resolving Images Australia (Melbourne, Australia). Anti-PAK4, were purchased from Cell Signaling Technology (Danvers, MA). Protein A-Agarose from Abcam (Melbourne, Australia), and [γ-P32]-adenosine triphosphate (γ-P32-ATP) from Perkin Elmer (Melbourne, Australia).
Kinase assay
Both purified recombinant PAK4 and PAK4 proteins immunoprecipitated (using anti-PAK4 antibody at 1:200) from PANC-1 cell lysates were subjected to a kinase assay. Both types of PAK4 were incubated with 2 μM cold ATP and 5 μg of myelin basic protein (MBP) in kinase buffer (40 mM HEPES pH 7.4, 10 mM MgCl2) on ice for 5 min before 10 μCi γ-P32-ATP was added and rotated at room temperature for 10 minutes. 2x SDS sample buffer was added to stop the reaction and the samples were then boiled at 95C for 5 minutes before loaded to a 12% SDS-PAGE. The gel containing separated proteins was fixed in 7% acetic acid at room temperature for 30 minutes, and then dried. The phosphorylated protein bands were developed and visualized through Fujifilm Bas-1800 II (Berthold, Australia) and the density of a band was calculated and analyzed using Multi Gauge (version 2.3) software.
Cell proliferation assay
Human and murine PC cells were incubated with different concentrations of PAKib in DMEM in the presence or absence of 5% FBS for 24 h. For the combination of PAKib with gemcitabine, the cells were pre-incubated with PAKib (10 μM) for 24h, followed by incubation with different concentrations of gemcitabine for another 24h. Cell proliferation was measured by MTT assay.
Measurements of cell death and cell cycle
Cell death and cell cycle were assayed using propidium iodide (PI) and flow cytometry. All floating and adhering cells were trypsinized and then washed two times with cold phosphate-buffered saline (PBS). For death assays, cells were harvested and resuspended in PBS containing 1 μg/mL PI. For cell cycle analysis, cells were harvested and resuspended in PBS containing 0.1% Triton X-100, 50 μg/mL PI, and 10 μg/mL RNaseA (Thermo Scientific), and then incubated at 37°C for 15 min. Samples were processed on with FACSymphony A3 flow cytometer (BD Biosciences, Melbourne, Australia) and the acquired data analysed with the FlowJo software (BD Biosciences, San Jose, CA).
Proteomic analysis
PANC-1 (1.25 × 106 cells) were seeded in a 10 cm culture dish, allowed to adhere for 2 h and then treated with 10 μM PAKib for 48 h and then lysed with radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris HCl, pH 8, 150 mM NaCl, 1% Triton x-100, 1% Na deoxycholate, 0.5% SDS, 1 mM EGTA, protease inhibitor, pHos sTop). The supernatant was precipitated with ice-cold acetone overnight at -20°C before resuspension in 8 M urea (in 50mM triethylammonium bicarbonate + 10mM TCEP) followed by incubation at 37°C for 30mins. Protein concentration was quantified using BCA protein assay and samples (containing equivalent amounts of total protein) were alkylated with 55 mM iodoacetamide for 45 mins at room temperature. The solution was diluted to a final concentration of 1M urea followed by overnight digestion at 37°C with sequencing-grade modified trypsin (Thermofisher) at a ratio of 1:50 (enzyme:protein). Digestion was stopped by addition of formic acid to a final concentration of 1% (v/v). The peptide mix was purified through solid phase extraction (SPE) with Oasis HLB cartridges (Waters) according to manufacturer's instructions. Eluted peptides were split into two tubes, Tube 1 (global mass spec) and Tube 2 (phospho-enrichment) and freeze-dried. Tube 1 was resuspended in 2% acetonitrile and 0.05% trifluoracetic acid before analysis by mass spectroscopy. Phosphorylated peptides from Tube 2 were purified using a titanium dioxide (TiO2) enrichment method. Peptides were mixed at a ratio of 1:6 (peptides: TiO2 beads) with TiO2 beads (GL sciences) in loading buffer (2M Lactic acid in 5% trifluoroacetic acid (TFA), 50% acetonitrile) and incubated for 1 hour at room temperature. The beads were washed twice with loading buffer and then three times with washing buffer (50% acetonitrile, 5% TFA). The phosphorylated peptides were eluted with 1% (v/v) ammonia solution (pH 11.3) and 30% (v/v) acetonitrile, and then acidified with 1μL formic acid per 10μL eluent. Eluted phosphorylated peptides were freeze-dried, resuspended with 2% acetonitrile and 0.05% TFA for mass spectroscopy. The data obtained from mass spectroscopy were processed through MaxQuant (version 1.6.17.0) [14] and search against the Homo Sapiens database (SwissProt Taxonomy ID 9606, updated Dec 2020). The search parameters are Trypsin as the cleavage enzyme and a maximum of 2 missed cleavages. Instrument parameters are based on the default Maxquant setup. Carbamidomethyl cysteine was set as fixed modification. The LFQ algorithm was used for label free quantitation and match between run was activated. Oxidation of methionine, acetylation of the protein N-terminus and phosphorylation of serine, threonine and tyrosine were considered as variable modifications. Protein and peptides groups were set to a maximum false discovery rate (FDR) of < 0.01. Statistical and network analysis of the search results was carried out before using Perseus software (version 1.6.14.0) [15] and Cytoscape (version 3.8.2) [16].
Mouse study
All mouse experiments were approved by the Austin Health Animal Ethics Committee (A2019/05654). Experimental mice were housed and cared in the BioResource Facility at Austin Health and monitored according to health criteria under the guideline of the Austin Health Animal Ethics Committee. Murine PC cells KPCPAK1WT833 (5 × 104 cells/100 μl culture medium) were subcutaneously injected into the flanks of 6-week-old, male C57BL6 mice. When a tumour reached 70 mm3 (in about 9 days) in set one experiment: control and PAKib, PAKib (40 mg/Kg) was given by subcutaneous injection (s.i.) every other day for two weeks. The control mice were given same volume of PEG400 (s.i.) which was used to dissolve PAKib. In set two experiment where mice were divided into three groups: control, gemcitabine alone, and gemcitabine plus PAKib, when a tumour reached 100 mm3, gemcitabine (25 mg/Kg) was given by intraperitoneal injection (i.p.) every fourth day, and PAKib was given by s.i. every other day in the group with combination treatment of gemcitabine plus PAKib. The control mice were given same volume of PEG400 (s.i.). The mice were treated for two weeks. Tumour growth was determined by tumour volume measured with a calliper every other day and by tumour weight measured at the end of each experiment.
Statistical analysis
All values are expressed as mean ± standard error. The in vitro data are from three independent experiments each in triplicate. The in vivo data were collated according to numbers of tumour samples. Data were analysed by one-way ANOVA or t-test (SPSS, IBM, New York, NY). Differences between two means with p < 0.05 were considered significant.
Results
PAKib inhibited the activity of PAK4
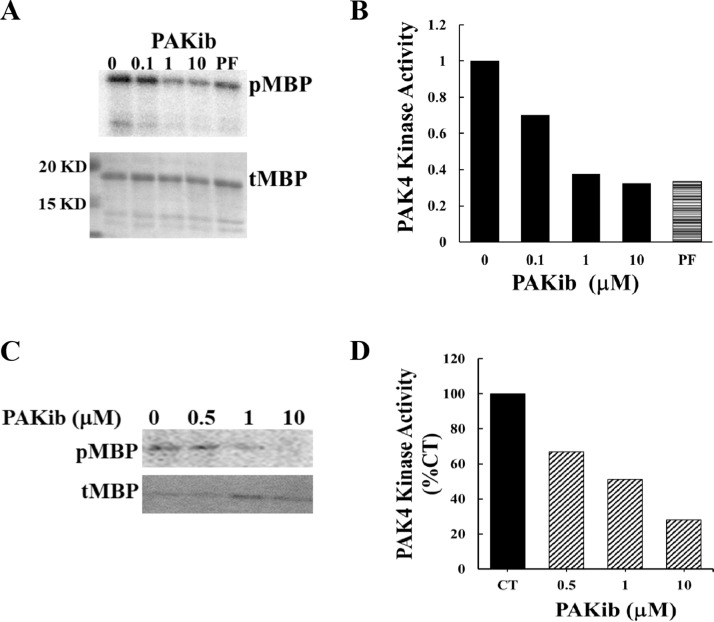
PAKib was synthetized and provided by PAKinax Pty. Ltd (Melbourne, Australia). PAKib suppressed the kinase activity of PAK4 with an IC50 value around 500 nM in an in vitro kinase assays using PAK4 immuno-precipitated from PC cells (Fig. 1A&B). PAKib also decreased the kinase activity of purified GST-PAK4 fusion protein (Fig. 1 C&D). The inhibition of PAKib on PAK4 was confirmed in a kinase assay using a Kinome panel containing about 400 kinases from Reaction Biology (Malvern, USA, data not shown). These results have demonstrated PAKib as a PAK4 inhibitor. Since PAK4 plays important role in PC development and progression, we have further investigated the role of PAKib in PC.
Fig. 1.
PAKib inhibited the kinase activity of PAK4. In an in vitro kinase assay using myelin basic protein (MBP) as substrate, PAKib suppressed the kinase activity of PAK4 (immune-precipitated from PANC-1 cell) with an IC50 value about 500 nM (A, B), and of a recombinant PAK4 (C, D). pMBP: phosphorylated MBP; tMBP: total MBP; CT: control. The density of pMBP from 0 (PAKib, B) or CT (D) were taken as 1 or 100% respectively.
PAKib inhibited PC cell growth and promoted the inhibition of gemcitabine on PC cells by inducing cell cycle arrest and cell death
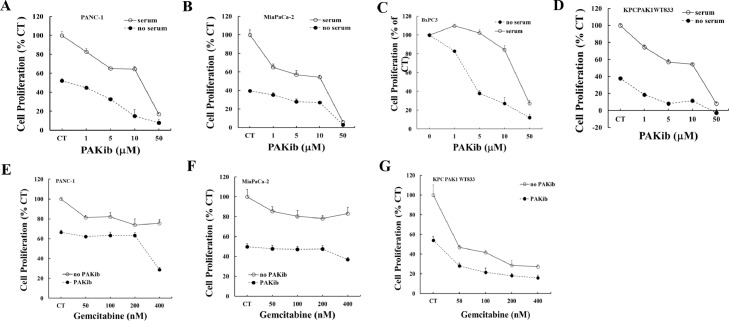
Gemcitabine has been considered as a standard treatment and widely used as a first-line drug in palliative treatment of pancreatic cancer [17]. On-going trials have been carried on testing the efficacy of different drugs, either alone or in combination with gemcitabine. Likewise, the effect of PAKib on cell proliferation with or without gemcitabine was determined by MTT assay. PAKib suppressed the proliferation of both human (PANC-1, MiaPaCa-2 and BxPC3) and murine (KPCPAK1WT833) PC cells (Fig. 2A-D, and Table 1) in the presence or absence of serum, with more potent inhibition in the absence of serum. PAKib decreased the proliferation of both KRas-mutated (PANC-1 and MiaPaCa-2) and KRas wild type (BxPC3) PC cells. Pre-treatment of PC cells with PAKib promoted the inhibitory effect of gemcitabine on PC cell by further reduction of proliferation (Fig. 2E-F).
Fig. 2.
PAKib decreased PC cell proliferation and enhanced the inhibitory effect of gemcitabine. Human PANC-1 (A), MiaPaCa-2 (B) and BxPC3 (C), and murine KPCPAK1WT833 (D) PC cell lines were treated with PAKib with concentrations indicated in the figures (A-D) with or without serum for 24h. The cell proliferation was determined by MTT assay as described in the Materials and Methods. PAKib dose-dependently decreased the proliferation of both human and murine PC cells. In the combination of PAKib and gemcitabine treatment, PANC-1 (E), MiaPaCa-2 (F) and KPCPAK1WT833 (G) were pre-treated with PAKib (10 μM) for 24h and followed by another 24h treatment of gemcitabine (Gem) with concentrations indicated in the figures (E-G). The date was summarized from three independent experiments. CT: control. The values from non-treated control were taken as 100%.
Table 1.
PAKib IC50 (mM) on cell proliferation
| PANC-1 | MiaPaCa-2 | BXPC3 | KPCPAKWT833 | |
|---|---|---|---|---|
| +serum | 13.77 ± 0.32 | 6.88 | 11.02 ± 0.89 | 9.49 ± 0.89 |
| -serum | 6.02 ± 0.23 | 9.63 ± 5.24 | 4.68 ± 0.20 | 1.23 ± 0.20 |
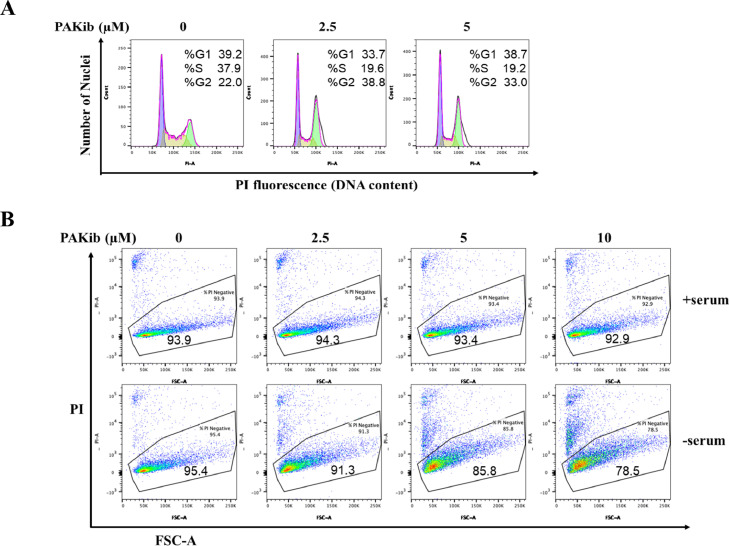
To determine the intracellular mechanism(s) of PAKib in its inhibition of PC cell growth, both human (PANC-1) and murine (KPCPAK1WT833) were treated with PAKib in the presence or absence of serum, changes in cell cycle and cell death were measured by PI staining followed by flow cytometry analysis. By 24 h treatment in serum, PAKib, from 2.5 μM, induced cell cycle arrest at G2/M phase in PANC-1 cells (Fig. 3A). In the absence of serum, PAKib caused cell death dose-dependently in KPCPAK1WT833 cells (Fig. 3B). These results indicated that PAKib inhibited PC cell growth by inducing cell cycle arrest (in human PC cells) or cell death (in murine PC cells).
Fig. 3.
PAKib suppressed PC cell growth by inducing cell cycle arrest and cell death. PANC-1 (A) and KPCPAK1WT833 (B) cells were treated with PAKib for 24h before being harvested for propidium iodide (PI) staining and flow cytometry as described in the Materials and Methods. The acquired data were analysed with the FlowJo software. The results were selected and represented from three independent experiments.
PAKib suppressed PC growth and enhanced the inhibitory effect of gemcitabine on PC in vivo
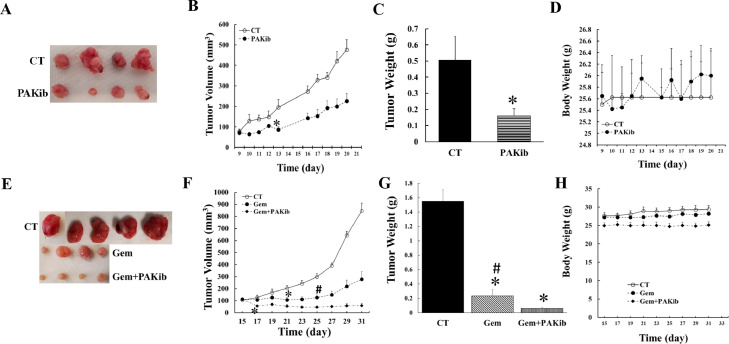
The effect of PAKib on pancreatic tumour growth was determined using a syngeneic mouse model where a murine PC cell line (KPCPAKWT833) was subcutaneously injected into the back of a hind leg of a mouse. PAKib (40 mg/Kg) was given by subcutaneous injection (s.i.) every second day when tumour volumes reached above 70 mm3 in one set of experiments (Fig. 4A-D). In another set of experiments, when tumour volumes were around 100 mm3, gemcitabine (25 mg/Kg) was given every fourth day by intraperitoneal injection (i.p.) alone or plus PAKib (40 mg/Kg, s.i. every second day) (Fig. 4E-H). PAKib suppressed tumour growth by reducing the average tumour volume to 47% of the control maximally at day 20 (Fig. 4B), and by decreasing the tumour weight to 32% of the control (Fig. 4C). No toxicity of PAKib was observed as shown by a stable mouse body weight (Fig. 4D).
Fig. 4.
PAKib suppressed PC growth in vivo and enhanced the inhibitory effect of gemcitabine on PC. KPCPAK1WT833 cells (50,000 cell/100 μl DMEM plus 5% FBS) were subcutaneously injected into the back of the right hind leg of a mouse. The mice were treated with PAKib (40 mg/Kg) by a subcutaneous injection (s.i.) every second day when tumour volume reached above 70 mm3 in set one (A-D) experiment where the initial average tumour volumes were 77.51±12.27 mm3 and 70.77±11.41 mm3 for control (CT, n=4) and PAKib (n=4) respectively (B). In set two experiment (E-H), when tumour volume reached 100 mm3, mice were treated with gemcitabine (Gem, 25 mg/Kg) alone every fourth day by intraperitoneal injection (i.p.) or Gem plus PAKic (40 mg/Kg, s.i. every second day). For set two experiment, the initial tumour volumes were 106.34+12.48 mm3, 113.12+13.82 mm3, and 114.53+3.94 mm3 for CT (n=5), Gem alone (n=4) and Gem plus PAKib (n=4) respectively (F). *, p<0.05 compared to CT; #, p<0.05 compared to Gem plus PAKib.
Gemcitabine significantly reduced the tumour volume from 12 days after starting treatment till the end of experiment, compared to control (Fig. 4F). Gemcitabine decreased the average tumour volume maximally to 33% (Fig. 4F) and the tumour weight to 15% (Fig. 4G) of the control respectively by the end of experiment. When compared to gemcitabine, the combined treatment of gemcitabine and PAKib further suppressed the tumour growth by reducing the tumour volume and weight to 21% (Fig. 4F) and 26% (Fig. 4G) of gemcitabine treated alone respectively. No significant toxicity was observed from the treatments of either gemcitabine alone or gemcitabine plus PAKib as shown by a stable mouse body weight (Fig. 4H). These results indicated that PAKib suppressed pancreatic tumour growth on its own and stimulated the inhibition by gemcitabine of PC growth in vivo.
PAKib inhibited PC via multiple singling pathways
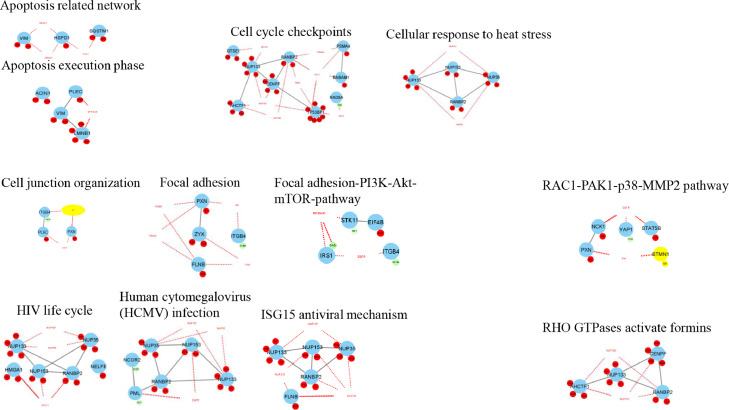
To discover the mechanism(s) involved in the effect of PAKib, PC cells were treated with PAKib for 48 h and then were lysed. The cell lysates were subjected to proteomic analysis as described in the Materials and Methods. There was no significant difference in the total protein abundances between control and PAKib-treated cells as shown from a global protein profile obtained from a mass spectrometry followed by a statistical analysis using Perseus (version 1.6.14.0). An unbiased phosphorylated peptide profile was also generated from the TiO2 enriched samples to determine changes in phosphorylated peptides and their related proteins. Upon statistical analysis using Perseus, a total of 285 phosphosites were significantly changed (Supplementary File 1) and they were mapped to 198 proteins (Fig. 5A). A heatmap was generated using the significant phosphosites after normalized by Z-score (Fig. 5B). 43 proteins out of 198 proteins with significant changes were identified in KEGG pathways. The phosphorylation of 16 proteins decreased by PAKib were identified in a distinctive pathway of KEGG (Table 2). The phosphorylation of 27 proteins were increased by PAKib and were involved in a distinctive pathway of KEGG (Table 3). Cell cycle, focal adhesion, tight junction, insulin-signaling, chemokine-signaling, MAPK-signaling, mTOR-signaling, p53-signaling, ErbB and Jak-STAT signaling pathways are among those KEGG pathways identified. The data with significant changes in phosphosites generated through Perseus was formatted and entered to Cytoscape [16] for analysis of interaction networks. 15 pathway networks were built up through a network analysis of Cytoscape (Fig. 6). Consistent with the findings from Perseus software analysis, cell cycle checkpoints, apoptosis, cell junction, focal adhesion, insulin signaling, and glucose metabolism networks were identified (Fig. 6). In addition, Rho-, Rac-activated pathways were identified. A few virus infection and anti-virus networks also showed up (Fig. 6).
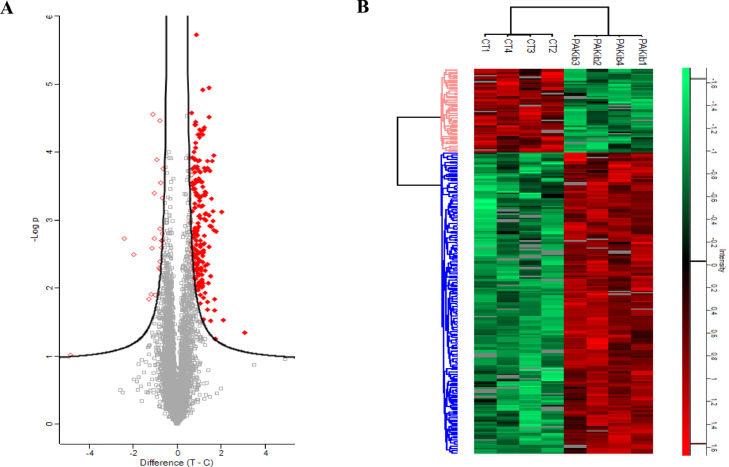
Fig. 5.
PAKib induced changes in phosphosites and the phosphorylated proteins. PANC-1 cells were treated with PAKib (10 μM) for 48h before the cells were lysed and the resultant cell lysates were subjected to a proteomic analysis as described in the Material and Methods. 4 repeated samples for control (CT) and PAKib-treated separately were applied in the experiment. The raw data generated through a Mass spectrum were processed using MaxQuant software, and resultant data were input into a Perseus software (version 1.6.14.0) where the changes in phosphosites and related phosphorylated proteins were identified. Statistically significant phosphosites were calculated and the mapped proteins were shown in the Volcano plot (A). The phosphosites with significant changes were used to generate a heatmap (B). Green represented the phosphorylated proteins decreased by PAKib compared to CT while Red PAKib-increased phosphorylated proteins. T: PAKib-treated; C: control
Table 2.
PAKib down-regulated phosphorylated proteins involved in KEGG pathways
| Gene names | KEGG pathways |
|---|---|
| ZAK | MAPK signaling pathway;Tight junction |
| PML | Acute myeloid leukemia;Endocytosis;Pathways in cancer;Ubiquitin mediated proteolysis |
| HMGCR | Bile secretion;Terpenoid backbone biosynthesis |
| PSD3 | Endocytosis |
| LMNA | Arrhythmogenic right ventricular cardiomyopathy (ARVC);Dilated cardiomyopathy;Hypertrophic cardiomyopathy (HCM) |
| ITGB4 | Arrhythmogenic right ventricular cardiomyopathy (ARVC);Dilated cardiomyopathy;ECM-receptor interaction;Focal adhesion;Hypertrophic cardiomyopathy (HCM);Regulation of actin cytoskeleton |
| ARHGEF11 | ko05152;Vascular smooth muscle contraction |
| STK11 | Adipocytokine signaling pathway;mTOR signaling pathway |
| UBE3A | Ubiquitin mediated proteolysis |
| ACLY | Carbon fixation pathways in prokaryotes;Citrate cycle (TCA cycle) |
| NCOR2 | Notch signaling pathway |
| IRS1 | Adipocytokine signaling pathway;Aldosterone-regulated sodium reabsorption;Insulin signaling pathway;Neurotrophin signaling pathway;Type II diabetes mellitus |
| SMC4 | Cell cycle - yeast |
| ACSS2 | Carbon fixation pathways in prokaryotes;Glycolysis / Gluconeogenesis;Methane metabolism;Propanoate metabolism;Pyruvate metabolism |
| LLGL2;LLGL1 | Tight junction |
| ABLIM1 | Axon guidance |
Table 3.
PAKib up-regulated phosphorylated proteins involved in KEGG pathways
| Gene names | KEGG pathways |
|---|---|
| MARCKS | Fc gamma R-mediated phagocytosis |
| NUP133 | RNA transport |
| NUP98 | RNA transport |
| RANBP2 | RNA transport |
| NUP35 | RNA transport |
| THOC2 | RNA transport;Spliceosome |
| UFD1L | Protein processing in endoplasmic reticulum |
| LMNA | Arrhythmogenic right ventricular cardiomyopathy (ARVC);Dilated cardiomyopathy;Hypertrophic cardiomyopathy (HCM) |
| PXN | Bacterial invasion of epithelial cells;Chemokine signaling pathway;Focal adhesion;Leukocyte transendothelial migration;Regulation of actin cytoskeleton;VEGF signaling pathway |
| STMN1 | MAPK signaling pathway |
| HSPD1 | ko05152;RNA degradation;Type I diabetes mellitus |
| ARFGAP1 | Endocytosis |
| ZYX | Focal adhesion |
| SQSTM1 | Osteoclast differentiation |
| RPL18 | Ribosome |
| UPP1 | Drug metabolism - other enzymes;Pyrimidine metabolism |
| WDR43 | Ribosome biogenesis in eukaryotes |
| GTSE1 | p53 signaling pathway |
| NCK1 | ErbB signaling pathway;Pathogenic Escherichia coli infection;T cell receptor signaling pathway |
| EIF4B | mTOR signaling pathway;RNA transport |
| COPA | Neuroactive ligand-receptor interaction |
| THOC1 | RNA transport;Spliceosome |
| FLNB | Focal adhesion;MAPK signaling pathway |
| PSMA4 | Proteasome |
| ACIN1 | mRNA surveillance pathway;RNA transport;Spliceosome |
| SF3B1 | Spliceosome |
| STAT5B | Acute myeloid leukemia; Chemokine signaling pathway; Chronic myeloid leukemia; ErbB signaling pathway; Jak-STAT signaling pathway;ko05152;Measles;Pathways in cancer |
Fig. 6.
PAKib affected intracellular pathways. The intracellular pathways affected by PAKib were calculated and identified using Cytoscape software. The data with significant changes in phosphosites generated through Perseus was formatted and entered to Cytoscape software where the related network pathways (solid grey lines) were identified, and the extended network pathways (red dotted lines) were also deduced. The red notes represented PAKib-increased phosphosites in related proteins while the green notes the PAKib-decreased phosphosites in related proteins.
In summary, PAKib could affect cell cycle and apoptosis via MARP-, ErbB-, mTOR-signaling pathways. PAKib could also regulate cell junction and focal adhesion via Rho- and Rac-dependent pathways. The involvement of PAKib in virus infection and anti-virus networks suggested a possible role of PAKib in inflammation and immune response.
Discussion
KRas mutations account for over 95% of PDA patients. KRas-driven cellular and animal cancer models have advanced our understanding of the initial and progression of various stages of PDA. However, there is very little progress in the development of KRas-targeted agents for the application in cancer treatment due to the absence of any druggable cavity in KRas protein structure. Recently specific inhibitors for mutant KRasG12C (glycine 12 to cysteine) have shown some effects on lung and colon cancers [18]. These inhibitors are not suitable for PDA as 90% of PDA carry KRasG12D (glycine 12 to aspartic acid) and KRasG12V (glycine 12 to valine) mutations [2], which are so far undruggable as the amino acids substituted by mutation are currently chemically unreactable [19].
PAKs, positioned at the junction of several oncogenic pathways [20] are key effectors of Rho family GTPases downstream of Ras. PAK4 is a key downstream effector of CDC42 [21]. PAK4 gene is amplified in PDA and possibly associated with poor prognosis [22]. Amplification of PAK4 is co-existed with oncogenic Ras in pancreatic cancer [23]. PAK4 expression level in PC cell lines is negatively related to gemcitabine sensitivity [24]. PAK4 also mediates pancreatic stellate cell-stimulated proliferation of PC cells by activating PI3K/Akt pathway [25].
We have demonstrated here that a newly developed PAK4 inhibitor, PAKib, suppressed PC growth in cell culture and in a syngeneic mouse model of PC, and that PAKib enhanced the inhibitory effect of gemcitabine on PC in vitro and in vivo. We have also revealed that PAKib inhibited PC cell growth by inducing cell death and cell cycle arrest, which is confirmed by our data obtained from the proteomic study showing the involvement of PAKib in cell cycle and apoptosis via multiple signaling pathways including MARP, ErbB, and mTOR.
The inhibition by PAKib of PC growth in vivo is quite potent, demonstrated by over 50% reduction of both tumour volume and weight by 10 days of treatment (Fig. 4B&C). PAKib also stimulated the inhibitory effect of gemcitabine on PC dramatically when combined with gemcitabine, by further decreasing tumour volume and weight to 21% and 27% of gemcitabine treatment alone respectively. These data provide a solid foundation for further testing this PAK4 inhibitor in clinical settings with PC patients. Except for PC cells, PAKib also suppressed growth of many other types of cancer cells including colorectal, lung, breast, brain, skin, prostate, and cholangiocarcinoma as shown in the supplementary Table 1, implicating the potential application of PAKib in the treatment of other types of cancers.
Recently PAK4 inhibition has been shown to improve PD-1 blockade immunotherapy in a skin cancer model of mice [26], indicating a role of PAK4 in tumour immune response. Most newly developed chemical compounds are initially tested in xenografted tumour models in Scid mice which have their immune system compromised. We have tested this novel PAK4 inhibitor, PAKib in a syngeneic mouse model in which the mice used have a fully functioning immune system. This enables us to explore the potential effect on tumour immune response. Indeed, our preliminary data from a flow cytometry study of tumour tissues dissected from mice treated by gemcitabine alone and gemcitabine plus PAKib, showed that PAKib may affect the infiltration of numbers of immune cells carrying markers of B cell (B220), T cell (CD4, CD8, PD-1) and myeloid cells (CD11b, Ly6c), and that PAKib may also affect the numbers of cells expressing PD-L1 (Supplementary Fig.1). These data implied the effect PAKib on the tumour immune response of PC. We are currently carrying on more extensive studies to investigate the role of PAKib in tumour immune response.
Only limited numbers of PAK4 inhibitors have been tested in human clinical trials although a few PAK4 inhibitors have been developed. PF-3758309, the first developed PAK4 specific inhibitor, failed in a phase I clinical trial (NCT00932126) because of unsatisfied pharmacokinetics (PK) and poor tumour response [27]. KPT-9274, a dual inhibitor targeting PAK4 and nicotin-amid phosphoribosyl transferase (NAMPT) is currently in a phase I clinical trial (NCT02702492) in patients with advanced solid tumours and non-Hodgkin lymphoma and its safety and efficacy are yet to be found out [28]. LCH-7749944, another PAK4 inhibitor showed inhibition of gastric cancer, but its clinical utility has yet to be explored [29]. Depside the preclinical agents developed, there is no product of PAK4-targeted compounds in clinic use. The unsuccessful clinical application of PAK4 inhibitors highlights the unmet clinical challenge of therapeutically targeting PAK4 in cancer treatment, specifically pancreatic cancer. PAKib, a newly developed PAK4 inhibitor suppressed PC in cells and in preclinical mouse models and enhanced the inhibitory effect of gemcitabine on PC in vitro and in vivo, which demonstrated a great potential of PAKib in clinical settings.
Funding
This work was supported by grants from Pancare Foundation, Austin Medical Research Foundation (HH-2021, MN-2020) and MDHS (Medicine Dental Health Science, University of Melbourne) Seeding Ideas Grants (MN2020, HH and CA 2021).
Author contribution
HH & MN made conceptual design of the study, performed mouse experiments and overall data analysis; HH & CD performed kinase assay and cell-based assays; CD and CA performed the mass spectroscopy; LD did the flow cytometry assay and analyzed the data; JZ was responsible for the design and production of the inhibitor; HH, CA and YM performed the analysis of the data obtained from the mass spectrum.
Declaration of Competing Interest
MN and JZ are on the Board of Pakinax Pty Ltd that was involved in the manufacture or the PAKib. JZ is current CEO of Pakinax Pty Ltd. HH is a medical advisor to Pakinax Pty Ltd and was involved in the manufacture of PAKib.
Acknowledgements
Dr. Hong He is supported by the Henry Baldwin Cancer Research Trust Fund.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101329.
Contributor Information
Hong He, Email: hong.he@unimelb.edu.au.
Mehrdad Nikfarjam, Email: m.nikfarjam@unimelb.edu.au.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, I Australian Pancreatic Cancer Genome. Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bournet B., Buscail C., Muscari F., Cordelier P., Buscail L. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities. Eur. J. Cancer. 2016;54:75–83. doi: 10.1016/j.ejca.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Wang K., Baldwin G.S., Nikfarjam M., He H. p21-activated kinase signalling in pancreatic cancer: New insights into tumour biology and immune modulation. World J. Gastroenterol. 2018;24:3709–3723. doi: 10.3748/wjg.v24.i33.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeo D., He H., Baldwin G.S., Nikfarjam M. The role of p21-activated kinases in pancreatic cancer. Pancreas. 2015;44:363–369. doi: 10.1097/MPA.0000000000000276. [DOI] [PubMed] [Google Scholar]
- 6.Thillai K., Sarker D., Wells C. PAK4 pathway as a potential therapeutic target in pancreatic cancer. Fut. Oncol. 2018;14:579–582. doi: 10.2217/fon-2017-0458. [DOI] [PubMed] [Google Scholar]
- 7.Wang K., Huynh N., Wang X., Baldwin G., Nikfarjam M., He H. Inhibition of p21 activated kinase enhances tumour immune response and sensitizes pancreatic cancer to gemcitabine. Int. J. Oncol. 2018;52:261–269. doi: 10.3892/ijo.2017.4193. [DOI] [PubMed] [Google Scholar]
- 8.Aboukameel A., Muqbil I., Senapedis W., Baloglu E., Landesman Y., Shacham S., Kauffman M., Philip P.A., Mohammad R.M., Azmi A.S. Novel p21-Activated Kinase 4 (PAK4) allosteric modulators overcome drug resistance and stemness in pancreatic ductal adenocarcinoma. Mol. Cancer Ther. 2017;16:76–87. doi: 10.1158/1535-7163.MCT-16-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K., Huynh N., Wang X., Pajic M., Parkin A., Man J., Baldwin G.S., Nikfarjam M., He H. PAK inhibition by PF-3758309 enhanced the sensitivity of multiple chemotherapeutic reagents in patient-derived pancreatic cancer cell lines. Am. J. Transl. Res. 2019;11:3353–3364. [PMC free article] [PubMed] [Google Scholar]
- 10.Yeo D., Huynh N., Beutler J.A., Christophi C., Shulkes A., Baldwin G.S., Nikfarjam M., He H. Glaucarubinone and gemcitabine synergistically reduce pancreatic cancer growth via down-regulation of P21-activated kinases. Cancer Lett. 2014;346:264–272. doi: 10.1016/j.canlet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeo D., He H., Patel O., Lowy A.M., Baldwin G.S., Nikfarjam M. FRAX597, a PAK1 inhibitor, synergistically reduces pancreatic cancer growth when combined with gemcitabine. BMC Cancer. 2016;16:24. doi: 10.1186/s12885-016-2057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K., Zhan Y., Huynh N., Dumesny C., Wang X., Asadi K., Herrmann D., Timpson P., Yang Y., Walsh K., Baldwin G.S., Nikfarjam M., He H. Inhibition of PAK1 suppresses pancreatic cancer by stimulation of anti-tumour immunity through down-regulation of PD-L1. Cancer Lett. 2020;472:8–18. doi: 10.1016/j.canlet.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 13.Ndubaku C.O., Crawford J.J., Drobnick J., Aliagas I., Campbell D., Dong P., Dornan L.M., Duron S., Epler J., Gazzard L., Heise C.E., Hoeflich K.P., Jakubiak D., La H., Lee W., Lin B., Lyssikatos J.P., Maksimoska J., Marmorstein R., Murray L.J., O'Brien T., Oh A., Ramaswamy S., Wang W., Zhao X., Zhong Y., Blackwood E., Rudolph J. Design of selective PAK1 inhibitor G-5555: improving properties by employing an unorthodox low-pK a polar moiety. ACS Med. Chem. Lett. 2015;6:1241–1246. doi: 10.1021/acsmedchemlett.5b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 15.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 16.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burris HA, 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo AM, Tarassoff P, Nelson R, Dorr FA, Stephens CD, Von Hoff DD. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 18.Nagasaka M., Li Y., Sukari A., Ou S.I., Al-Hallak M.N., Azmi A.S. KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer Treat. Rev. 2020;84 doi: 10.1016/j.ctrv.2020.101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang C.V., Reddy E.P., Shokat K.M., Soucek L. Drugging the 'undruggable' cancer targets. Nat. Rev. Cancer. 2017;17:502–508. doi: 10.1038/nrc.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radu M., Semenova G., Kosoff R., Chernoff J. PAK signalling during the development and progression of cancer. Nat. Rev. Cancer. 2014;14:13–25. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakoshima T., Shimizu T., Maesaki R. Structural basis of the Rho GTPase signaling. J. Biochem. 2003;134:327–331. doi: 10.1093/jb/mvg149. [DOI] [PubMed] [Google Scholar]
- 22.Kimmelman A.C., Hezel A.F., Aguirre A.J., Zheng H., Paik J.H., Ying H., Chu G.C., Zhang J.X., Sahin E., Yeo G., Ponugoti A., Nabioullin R., Deroo S., Yang S., Wang X., McGrath J.P., Protopopova M., Ivanova E., Zhang J., Feng B., Tsao M.S., Redston M., Protopopov A., Xiao Y., Futreal P.A., Hahn W.C., Klimstra D.S., Chin L., DePinho R.A. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19372–19377. doi: 10.1073/pnas.0809966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S., Auletta T., Dovirak O., Hutter C., Kuntz K., El-ftesi S., Kendall J., Han H., Von Hoff D.D., Ashfaq R., Maitra A., Iacobuzio-Donahue C.A., Hruban R.H., Lucito R. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol. Ther. 2008;7:1793–1802. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moon S.U., Kim J.W., Sung J.H., Kang M.H., Kim S.H., Chang H., Lee J.O., Kim Y.J., Lee K.W., Kim J.H., Bang S.M., Lee J.S. p21-activated kinase 4 (PAK4) as a predictive marker of gemcitabine sensitivity in pancreatic cancer cell lines. Cancer Res. Treat. 2015;47:501–508. doi: 10.4143/crt.2014.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M., Guo H., Wang Q., Chen K., Marko K., Tian X., Yang Y. Pancreatic stellate cells derived exosomal miR-5703 promotes pancreatic cancer by downregulating CMTM4 and activating PI3K/Akt pathway. Cancer Lett. 2020;490:20–30. doi: 10.1016/j.canlet.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Abril-Rodriguez G., Torrejon D.Y., Liu W., Zaretsky J.M., Nowicki T.S., Tsoi J., Puig-Saus C., Baselga-Carretero I., Medina E., Quist M.J., Garcia A.J., Senapedis W., Baloglu E., Kalbasi A., Cheung-Lau G., Berent-Maoz B., Comin-Anduix B., Hu-Lieskovan S., Wang C.Y., Grasso C.S., Ribas A. PAK4 inhibition improves PD-1 blockade immunotherapy. Nat. Cancer. 2020;1:46–58. doi: 10.1038/s43018-019-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Song P., Gao Y., Shen L., Xu H., Wang J., Cheng M. Drug discovery targeting p21-activated kinase 4 (PAK4): a patent review. Expert Opin. Ther. Pat. 2021:1–11. doi: 10.1080/13543776.2021.1944100. [DOI] [PubMed] [Google Scholar]
- 28.Mpilla G.B., Uddin M.H., Al-Hallak M.N., Aboukameel A., Li Y., Kim S.H., Beydoun R., Dyson G., Baloglu E., Senapedis W.T., Landesman Y., Wagner K.U., Viola N.T., El-Rayes B.F., Philip P.A., Mohammad R.M., Azmi A.S. PAK4-NAMPT dual inhibition sensitizes pancreatic neuroendocrine tumors to everolimus. Mol. Cancer Ther. 2021 doi: 10.1158/1535-7163.MCT-20-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Wang J., Guo Q., Wang Y., Zhou Y., Peng H., Cheng M., Zhao D., Li F. LCH-7749944, a novel and potent p21-activated kinase 4 inhibitor, suppresses proliferation and invasion in human gastric cancer cells. Cancer Lett. 2012;317:24–32. doi: 10.1016/j.canlet.2011.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.