Figure 2. Variation Window Redesigned to include HGVS annotations.
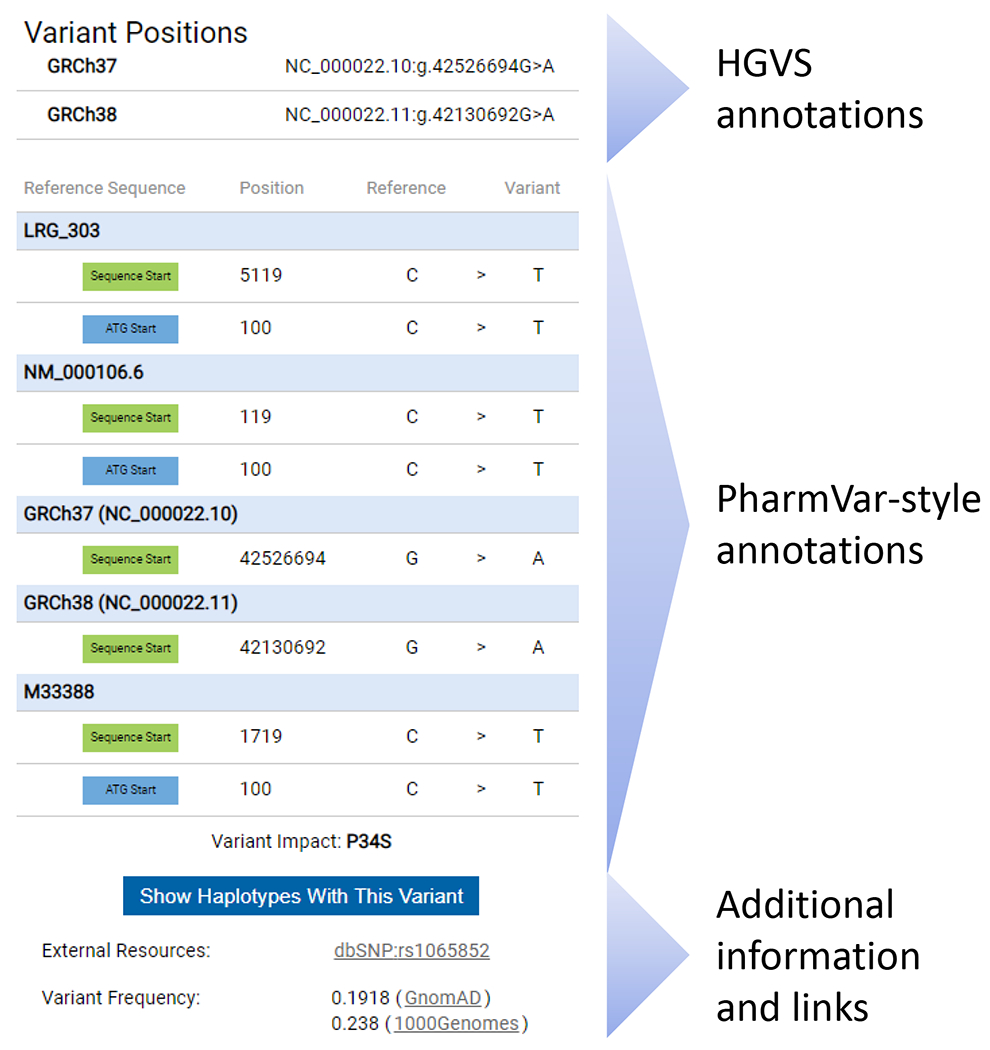
The new Variation Window is exemplified for a variant that is present on numerous CYP2D6 haplotypes, or star alleles. The Variation Window is activated by clicking on a SNV on a gene page. The top portion of the Variation Window displays SNV coordinates according to HGVS nomenclature on the gene, transcript and genome (GRCh37 and GRCh38) levels. Coordinates are displayed as obtained through the NCBI Variation Services and are only manually curated if no results are returned. The middle portion displays SNV positions ‘PharmVar-style’ on the gene, transcript and genome (GRCh37 and GRCh38) levels listing positions for both count modes (sequence start and the ‘A’ of the ATG’ start codon being +1). Note that PharmVar consistently uses the 3’ Rule to determine the positions of inserted or deleted nucleotide(s) and thus, HGVS and ‘PharmVar-style’ positions may differ for insertion/deletion variants in some instances which is most likely explained by differences in sequence alignments. PharmVar also displays single nucleotide insertions as ‘ins’ while HGVS displays them as duplications or ‘dup’. Additional details and examples are provided in the PharmVar ‘Standards document’ (4). The bottom portion of the variation window provides a filter option to display all variants with the selected SNV, the link to dbSNP (if a rsID exists), as well as SNV frequency information. There is also a bar providing the option to display all haplotypes with the selected variant.
