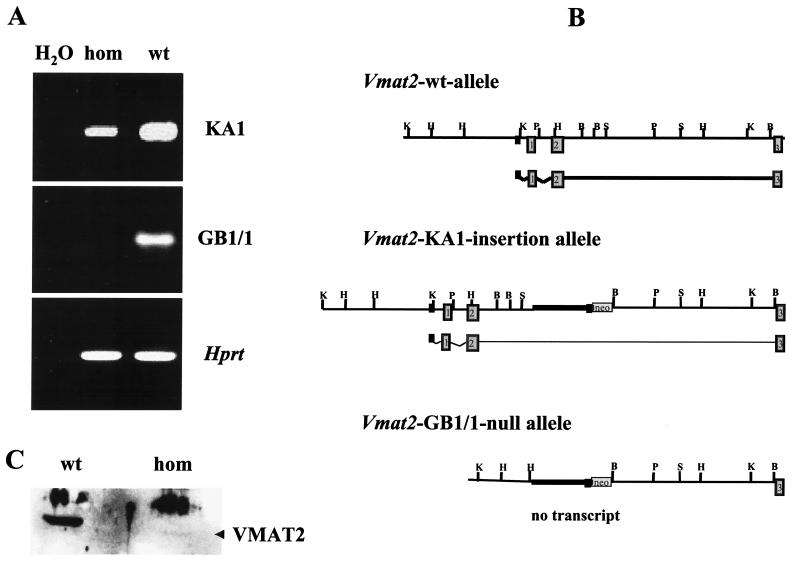
FIG. 3.
RT-PCR and Western blot analysis of homozygous KA1 insertional mutants and homozygous GB1/1 knockout mice. (A) Ethidium-stained agarose electrophoresis of RT-PCR products. Total midbrain RNA from homozygous (hom) and wild-type (wt) GB1/1 and KA1 neonates was isolated and reverse transcribed. Expression of Vmat2 mRNA was detected using specific primers for exon 2 and exon 12 of the mouse Vmat2 gene (33). These primers amplified Vmat2 cDNA made from homozygous KA1 mice, but no amplification product was generated with cDNA made from homozygous GB1/1 neonates. The quality of all cDNA preparations was examined by performing control RT-PCRs with primers to hypoxanthine phosphoribosyltransferase (Hprt) (22). (B) Schematic representation of the genomic organization of the Vmat2 wild-type allele, the KA1 insertion allele, and the GB1/1 knockout allele. The wild-type allele is transcriptionally active. The KA1 insertion interferes severely with transcription, and only a small amount of Vmat2 message is generated. In the GB1/1 knockout line, the first and second exon of the Vmat2 gene are deleted, completely abolishing the generation of normal Vmat2 message. (C) Western blot of striatal membrane preparations from homozygous and wild-type KA1 mice probed with a polyclonal antibody against the VMAT2 protein. Both lanes contain comparable amounts of protein, as determined by the Bradford method. Quantitative analysis of the blot using SeeScan indicated a decrease in signal of more than 95% in homozygotes below that of the wild-type signal. The position of the VMAT2-specific band is indicated by the arrowhead. The higher molecular weight band is nonspecific, being found in extracts from controls and mutants.