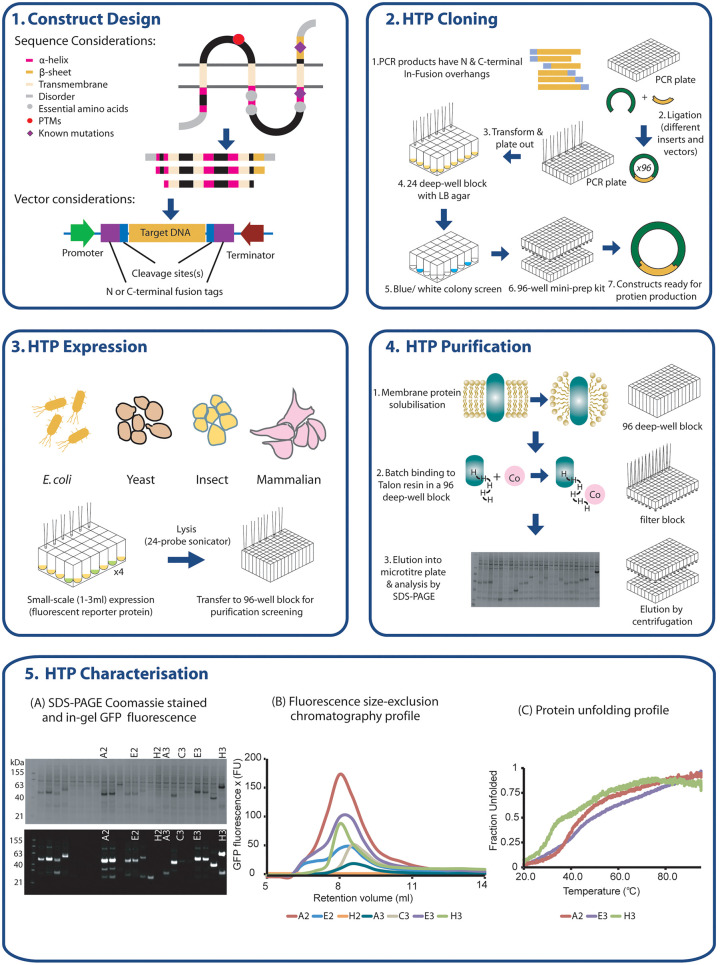
Figure 1. An illustrative overview of the HTP production of membrane proteins.
(Step 1) Bioinformatics servers and databases such as PSIpred [7] and Uniprot [8] are used to aid construct design, highlighting secondary structural features, domain boundaries critical residues, PTMs and mutant and target isoforms. six–24 constructs are designed for each MP target. Constructs include truncations, (of mainly disordered regions and domain boundaries), functional and disease mutations, and a variety of fusion-tags. (Step 2) HTP-cloning enables a diverse library of clones to be established in a few days. (Step 3) HTP expression enables the parallel production of all cloned constructs in a few days to 2 weeks depending on expression system. (Step 4) HTP purification using 96-well blocks and filter plates to purify 96 different conditions (constructs, encapsulation reagents, solvent conditions or additives) in under 12 h. (Step 5) HTP characterisation using (A) SDS–PAGE to assess protein purity, yield and susceptibility to proteases. Protein bands can be excised and analysed using mass spectrometry to identify the target protein or contaminants. Here 24 constructs are shown. Strong protein bands are observed for A2, E2 and H3 among others. Protein bands must usually be observable after Coomassie staining as well as GFP fluorescence. For example, although A3 can be detected by GFP florescence, the expected yield would be too low to make this construct tractable. Some degradation is observed for multiple constructs (not uncommon when using a GFP fusion). Note the high sensitivity of GFP. (B) F-SEC enables an assessment of monodispersity and provisional oligomeric state. Well-behaved MPs tend to have a clear monodisperse profile but this is not an indication of long-term stability. Seven constructs are labelled here and correspond to bands on the SDS–PAGE gel. GFP is not essential for this analysis. We also use tryptophan fluorescence for MPs lacking GFP tags, especially when GFP removal is destabilising. In these cases twin-strep tags are used to ensure the higher purity required to interpret the F-SEC profiles (C) Nano-DSF enables the temperature at which half of a protein sample is folded to be determined and is a good estimate of long-term stability. Ideally, a Tm½ would be more than 40°C. Not all MPs are shown here as low-yielding constructs will not be detected by nano-DSF. A2 and E3 are the most encouraging constructs with good SEC profiles and Tm½ values. Although H3 has a good SEC-profile its thermostability is lower (∼30°C) suggesting that the purification conditions require optimisation.