Figure 3.
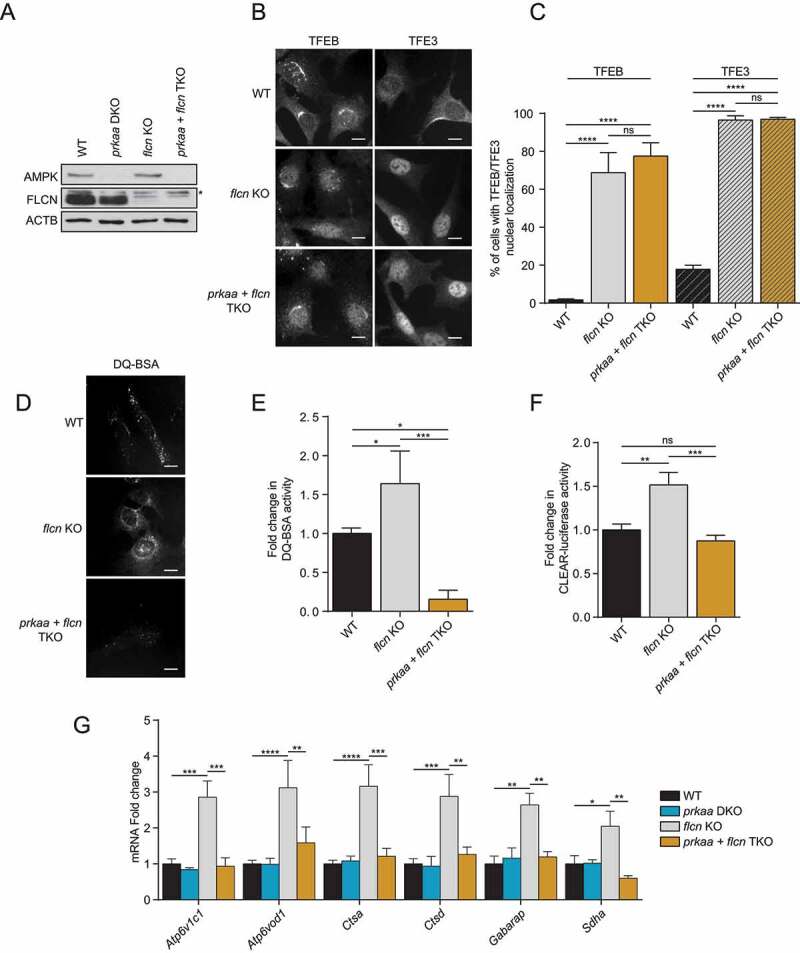
AMPK is required for FLCN-dependent activation of TFEB and TFE3. (A) Immunoblot of protein lysates of MEFs with indicated genotypes were incubated in complete media. Data are representative of three independent experiments. * represents nonspecific bands. (B) Representative images of indicated MEF lines incubated in complete media. Cells were fixed, permeabilized, and stained with an antibody against TFEB or TFE3. Scale bar: 20 μm. Images are representative of three independent experiments. (C) Quantification of the percentage of nuclear TFEB or TFE3 in MEFs incubated in complete media (mean ± SEM of three independent experiments, one‐way ANOVA, ns = not significant, ****P < 0.0001; n > 200 cells per condition). (D) Representative images of indicated MEF lines after 1 h of incubation with DQ-BSA-Red followed by a 2 h chase in complete media prior to fixation. Scale bar: 20 μm. Images are representative of three independent experiments. (E) Relative lysosomal activity, as determined by DQ-BSA assay in MEFs upon treatment as indicated in (D) (mean ± SEM of three independent experiments, one‐way ANOVA, *P < 0.05, ***P < 0.001; n > 200 cells per condition). (F) Relative Tfeb and Tfe3 transcriptional activity, as determined by CLEAR-luciferase promoter activity normalized against CMV-Renilla in MEFs incubated in complete media (mean ± SEM of the luminescence fold change from six independent experiments, one-way ANOVA, ns = not significant, **P < 0.01, ***P < 0.001) (G) Relative quantitative real‐time PCR analysis of Tfeb and Tfe3 target genes mRNA transcript levels in MEFs incubated in complete media (mean ± SEM of the RNA fold change of indicated mRNAs from four independent experiments, two-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
