Figure 7.
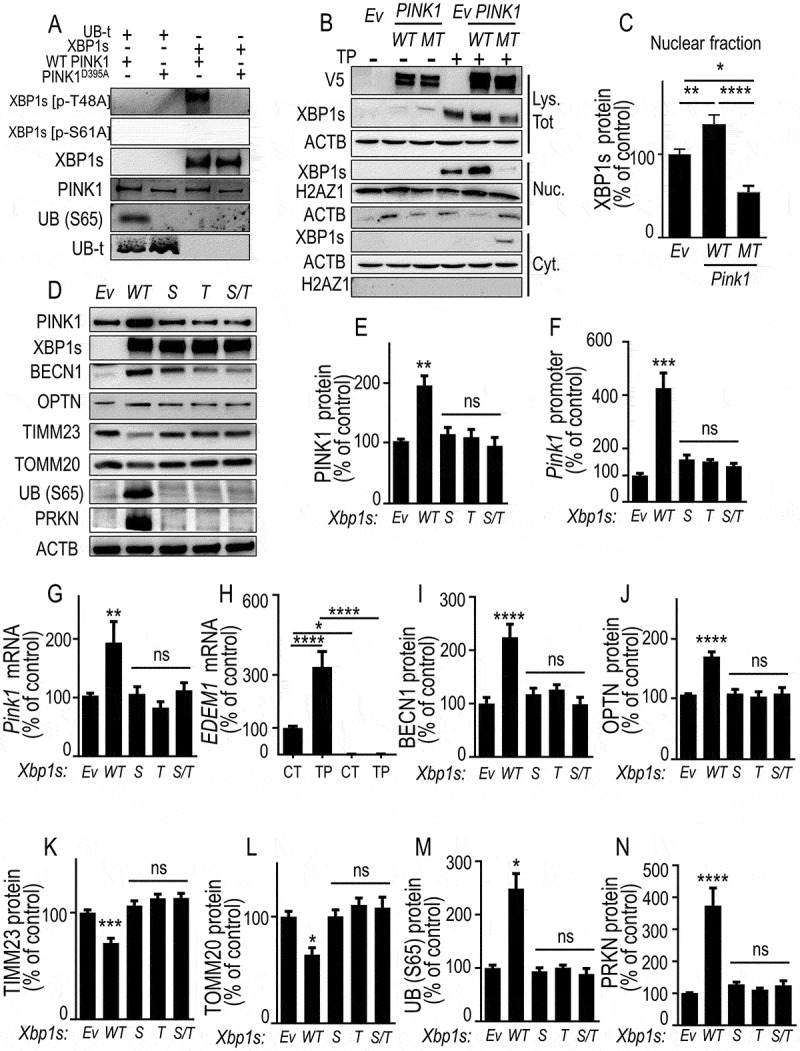
PINK1-mediated phosphorylation of XBP1s enhances its nuclear translocation, controls its own transcription and mitophagy, a phenotype abolished by non-phosphorylable XBP1s mutants. (A) XBP1s and UB (S65) (used as positive control substrate) phosphorylations by recombinant wild-type PINK1 (WT PINK1) or kinase-dead mutant PINK1 (PINK1D395A) were performed as described in Methods. XBP1s, XBP1s [p-T48A], XBP1s [p-S61A], total UBB (UB-t), UB (S65) and PINK1 protein levels were analyzed by western blot. (B) SH-SY5Y cells were transfected with empty vector (Ev), WT PINK1 or PINK1D395A (MT) cDNA. Twenty-four hours after transfection, cells were treated for 8 h without (-) or with (+) thapsigargin (TP, 1 µM). XBP1s, V5-tagged PINK1 and ACTB protein expressions were monitored in whole cell lysate (Lys. Tot.) while XBP1s, histone (H2AZ1) and ACTB protein expressions were monitored by western blot in either cytoplasmic (Cyt.) or nuclear (Nuc.) cellular fractions prepared as described in Methods. (C) Data corresponding to nuclear XBP1s are expressed as percent of Ev-untreated cells (taken as 100%) and are the means ± SEM of 2 independent experiments performed in triplicate. Statistical significances were analyzed by one-way ANOVA, Tukey’s multiple comparisons test * P < 0.05, ** P < 0.01, **** P < 0.0001. (D-G) SH-SY5Y were transiently transfected with an empty vector (Ev), wild-type Xbp1s (WT) or Xbp1s mutants (Xbp1s [p-S61A] (S), Xbp1s [p-T48A] [T] or Xbp1s [p-S61A]/Xbp1s [p-T48A] [S/T]) cDNAs. Twenty-four hours after transfection, PINK1 protein expressions (D and E, N = 12), promoter activity (F, N = 12), and mRNA levels (G) were analyzed as described in Methods. Data are expressed as percent of control Ev-transfected cells (taken as 100%) and are the means ± SEM of 4 independent experiments performed in triplicates. Statistical significances were analyzed by one-way ANOVA, Turkey’s multiple comparisons test, ns, non-significant, ** P < 0.01, *** P < 0.01. (H) EDEM1 mRNA was measured in basal (CT) or thapsigargin (TP)-stimulated conditions in wild-type (PINK1 CT, black bars) or PINK1 KD SH-SY5Y cells (note that EDEM1 mRNA are totally undetectable in PINK1 KD cells). Data are expressed as percent of control (CT)-untreated cells (taken as 100%) and are the mean ± SEM of 4 independent experiments performed in triplicates. Statistical analysis was performed by two-way ANOVA, Sidak’s multiple comparison test, * P < 0.05, **** P < 0.0001. (D, I-N) Expressions of BECN1 (D and I), OPTN (D and J), TIMM23 (D and K), TOMM20 (D and L) UB (S65) (D and M) and PRKN (D and N) were analyzed by western blotting after Ev-, WT Xbp1s or S, T and S/T Xbp1s-mutants cDNAs transfection as described in Methods. Data are expressed as percent of Ev cells (taken as 100%) and are the mean ± SEM of 3 independent experiments performed in triplicates. Statistical analysis were analyzed by one-way ANOVA followed by either Kruskal-Wallis multiple comparison test (E,L,M) or Tukey’s multiple comparison (F,G,I-K,N), * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001 and ns = non-significant
