Figure 2.
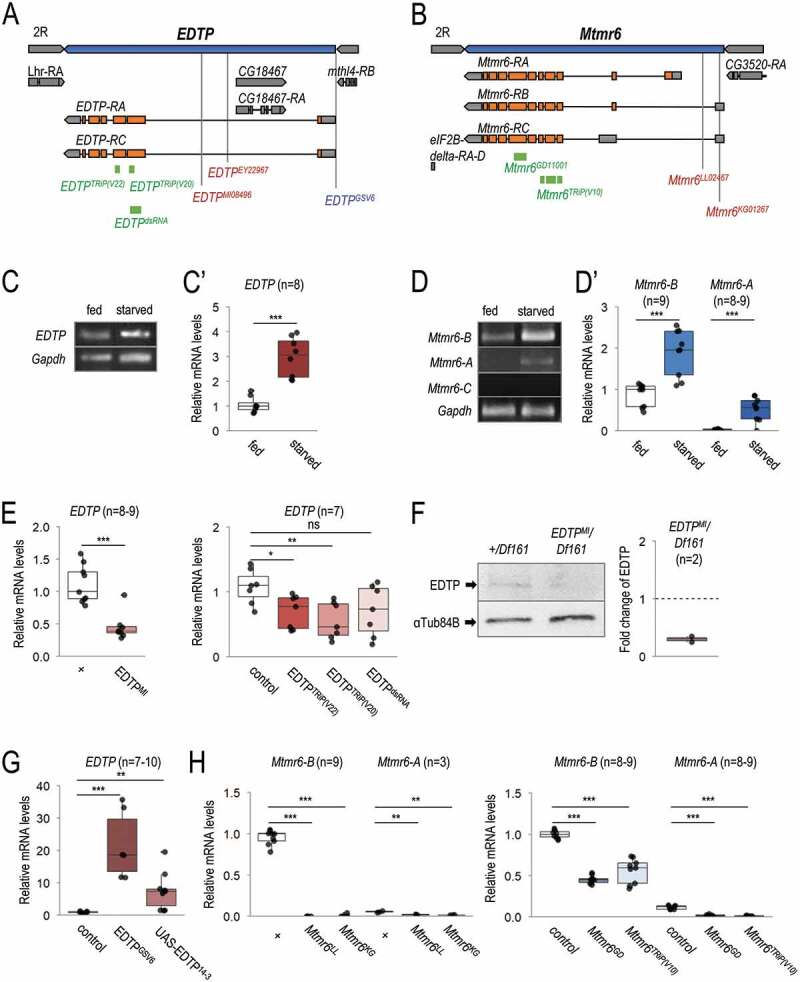
Mutant alleles, RNAi and overexpression constructs of EDTP and Mtmr6. (A) Exon/intron structure of EDTP. Specific positions of on-target sequences of RNAi constructs (green), inactivating mutations (red) and overexpressing insertion (blue) are shown. (B) Genomic structure of Mtmr6/CG3530. RNAi constructs (green) and mutant alleles (red) are indicated. (C-C’) EDTP is expressed in larval fat body cells of well-fed larvae, and its expression becomes elevated upon amino acid starvation, based on semi-quantitative RT-PCR (C) and quantitative real-time (qRT-) PCR (C’) analyses. (D) Splice-variants of Mtmr6 are expressed at different levels in the larval fat body. Under nutrient-rich condition, only Mtmr6-B is detectable by semi-qRT-PCR, the level of which becomes elevated in response to amino acid starvation. The expression of Mtmr6-A can be observed only under the latter condition. Mtmr6-C expression is not detectable under either circumstance. (D’) Based on qRT-PCR results, Mtmr6-B is markedly expressed, while Mtmr6-A shows a very weak expression in well-fed larvae. Expression of both splice variants is elevated upon starvation. (E) Relative transcript levels of the EDTPMI gene trap mutant allele and EDTP RNAi constructs, determined by qRT-PCR. (F) EDTPMI gene trap mutant allele also effectively lowers EDTP protein level. (G) Relative EDTP mRNA level is increased upon applying overexpression constructs. (H) Expression levels of active Mtmr6 splice variants (A and B) in control, mutant and RNAi-treated samples. qRT-PCR and semi-qRT-PCR were used on total RNA samples isolated from larval fat bodies of well-fed (C-D’) or 3 h-starved (C-H) animals at the third instar feeding larval (L3F) stage. Protein samples also stem from larval fat bodies of 3 h-starved animals at the third instar feeding larval (L3F) stage. Gapdh and αTub84B were used as internal controls for PCR experiments and in western blot analysis, respectively. Expression of UAS constructs was driven by Act5C-Gal4. In the case of long hairpin RNAi constructs, UAS-Dcr-2 was also coexpressed. w1118 animals (indicated by “+”) served as control for mutant strains, Act5C-Gal4/+ animals did for short hairpin RNAi and overexpression constructs, and UAS-Dcr-2/+; Act5C-Gal4/+ animals did for long hairpin RNAi constructs. Quantifications of normalized mRNA and protein levels are shown in box plots. *: p < 0.05, **: p < 0.01 ***: p < 0.001, ns: not significant. For statistics, see the Materials and Methods section
