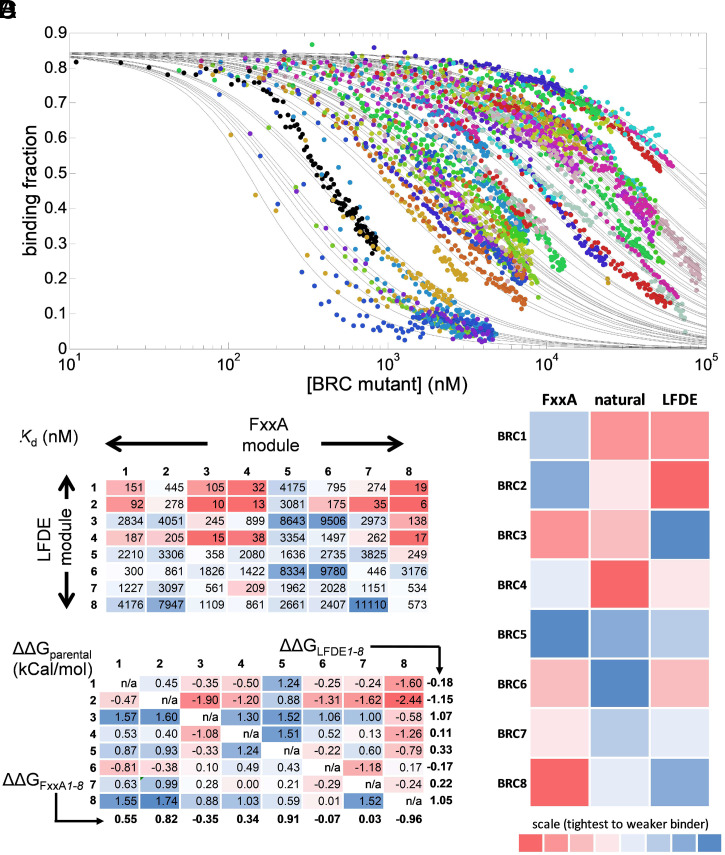
Fig. 2.
Affinity determination of 64 chimeric BRC4 repeats by a microfluidic droplet-on-demand system interfaced with fluorescence anisotropy detection. (A) Fraction of BRC4fl peptide bound to monomeric RAD51 as a function of GB1-BRC peptide chimera concentration for nine examples of the dataset of 64 combinations, measured using the droplet-on-demand anisotropy competition assay. Measurement conditions were 100 nM BRC4fl, 150 nM monomeric RAD51 in a buffer of 20 mM CHES (pH 9.5), 100 mM NaCl, 1 mM EDTA, at 20 °C. Note the starting binding fraction of 0.85 that is calculated from the affinity of the BRC4fl peptide for monomeric RAD51 and the initial concentrations used (50). (All 64 binding curves are shown separately elsewhere (SI Appendix, Fig. S7). (B) Kd values (in nM) determined for all 64 BRCA2 peptide chimeras using data in A. (C) Analysis of the effect of recombination, expressed as the difference in ΔG (SI Appendix, Table S4) of each shuffled variant relative to the average of the two natural parental combinations (ΔΔGparental, in kCal/mol) for each variant. The parental combinations are depicted in gray (by definition, their ΔΔGparental is always zero). The values indicated below each column and next to each row represent the average ΔΔGparental value for FxxA and LFDE modules from each repeat, respectively, and are referred to as ΔΔGFxxA1–8 and ΔΔGLFDE1–8, respectively. (D) Binding rank order of the parental BRC repeats (Central column) and the individual FxxA (Left column) and LFDE (Right column) modules comprising the repeats, indicated by intensity (low intensity = top binder, high intensity = poor binder, as indicated by scale bar). The contributions to binding of the eight different repeat-derived modules are calculated using their ΔΔGLFDE1–8 and ΔΔGFxxA1–8 values.