Abstract
To ensure genome integrity, the joining of breaks in the phosphodiester backbone of duplex DNA is required during DNA replication and to complete the repair of almost all types of DNA damage. In human cells, this task is accomplished by DNA ligases encoded by three genes, LIG1, LIG3 and LIG4. Mutations in LIG1 and LIG4 have been identified as the causative factor in two inherited immunodeficiency syndromes. Moreover, there is emerging evidence that DNA ligases may be good targets for the development of novel anti-cancer agents. In this graphical review, we provide an overview of the roles of the DNA ligases encoded by the three human LIG genes in DNA replication and repair.
Introduction
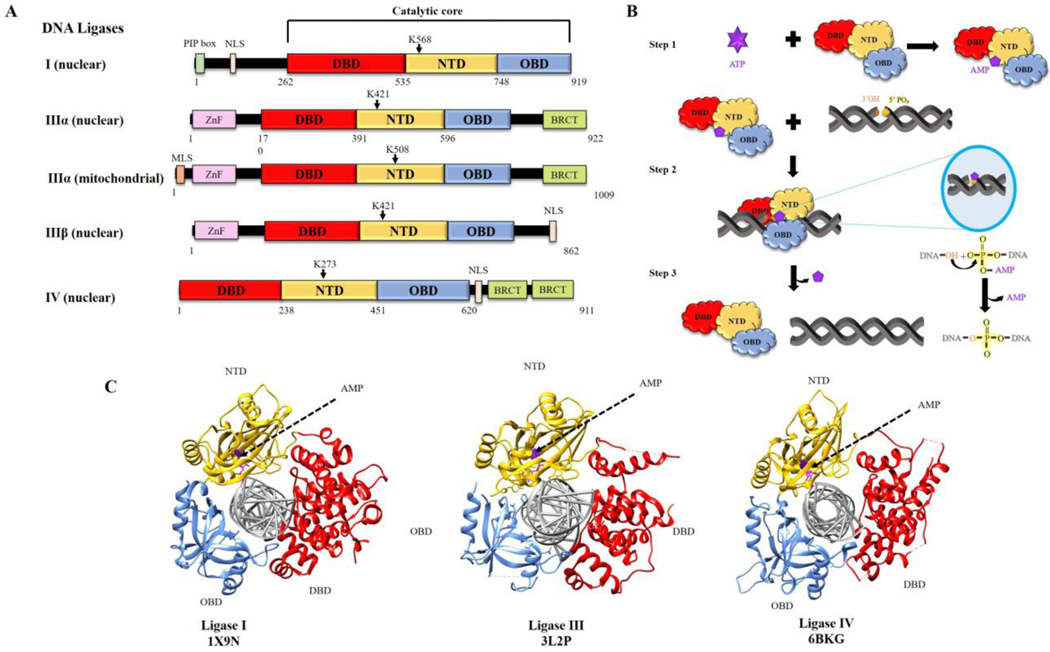
While biochemical studies suggested that mammalian cells contained multiple species of DNA ligase, this remained controversial for many years until the cloning of three human genes encoding DNA ligases (1). Notably, alternative translation initiation and alternative splicing mechanisms generate three distinct DNA ligase polypeptides from the LIG3 gene (1,2) in addition to the single DNA ligase polypeptides encoded by the LIG1 and LIG4 genes (Fig. 1A). The human DNA ligases share a related catalytic region composed of three domains, a DNA Binding Domain (DBD), a Nucleotidyl Transferase Domain (NTD) and an Oligonucleotide/Oligosaccharide-Binding-fold Domain (OBD) (Fig. 1A). In contrast to the high degree of amino acid sequence identity shared by the NTDs and OBDs that form the catalytic core, the amino acid sequences of the DBDs are more divergent (1).
Fig. 1. DNA ligases encoded by the three human LIG genes; ligation reaction mechanism and structure of catalytic regions complexed with nicked DNA.
A. Human DNA ligases (I, IIII and IV) share a conserved catalytic region comprised of DNA binding domain (DBD, red), nucleotidyl transferase domain containing the lysine residues that form the phosphoramidite bond with AMP (NTD, yellow) and oligonucleotide binding or OB-fold containing domain (OBD, blue). PCNA interacting peptide box (PIP box, light green), Zinc finger DNA binding domain (Znf, pink), BRCA1 C terminus domain (BRCT, lime green), mitochondrial localization signal (MLS, brown) and nuclear localization signal (NLS, white) are indicated. B. Schematic diagram of ligation reaction; step 1: Formation of ligase-adenylate using ATP. Step 2: Ligase-adenylate engages nicked DNA and transfers AMP to 5’phospahe termini, forming DNA-adenylate. Step 3: Non-adenylated Ligase interacts with DNA-adenylate to form phosphodiester bond. C. Crystal structures of DNA ligase catalytic regions with nicked DNA. Images were generated using the software Chimera (http://www.cgl.ucsf.edu/chimera) (23) and structures from RCSB PDB.
The human DNA ligases are members of the nucleotidyl transferase family that also includes mRNA capping enzymes and RNA ligases (3,4). Nucleotidyl transferases contain a conserved NTD-OBD catalytic core that interacts with a nucleotide co-factor to form a covalent enzyme-nucleoside monophosphate intermediate in which the nucleoside monophosphate is linked via a phosphoramidite bond to a specific lysine residue within the NTD. Unlike the NAD-dependent E coli DNA ligase, bacteriophage and human DNA ligases utilize ATP as their co-factor (Fig. 1B). While the three catalytic domains are arranged in an open formation during the formation of the DNA ligase-adenylate intermediate, they adopt a C-clamp structure when they engage with adjacent 3’ hydroxyl and 5’ phosphate termini that can occur either at single strand nicks in duplex DNA or juxtaposed duplex DNA ends. It is within this structure that the transfer of the AMP moiety to the 5’ phosphate termini, occurs forming the DNA-adenylate. Despite the divergence of amino acid sequence between the DBDs, the structures of the catalytic regions of the three human DNA ligases complexed with nicked DNA are remarkably similar with the three domains encircling and contacting the nicked DNA (5) (Fig. 1C). Finally, the non-adenylated DNA ligase engages with the DNA–adenylate and, utilizing the hydroxyl group at the 3’ termini as a nucleophile, catalyzes the formation of a phosphodiester bond and the release of AMP (Fig. 1B).
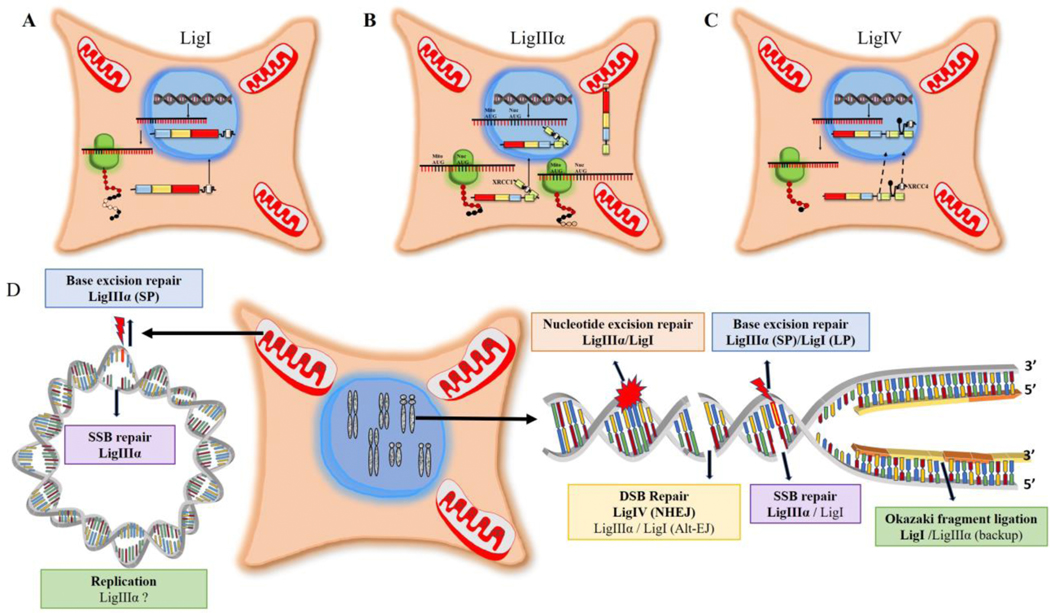
DNA ligases I (LigI) and IV (LigIV) function exclusively in the nucleus whereas DNA ligase III (LigIII) functions in both in the nucleus and mitochondria. LigI has a classic nuclear localization signal (NLS, residues 119–131, Fig. 1A) within its non-catalytic N-terminal region that targets this enzyme to the nucleus (6) (Fig. 2A). Similarly, LigIV contains a NLS (residues 623–638, Fig. 1A) within its non-catalytic C-terminal region. However, while it has been reported that the LigIV NLS is required for the nuclear localization of both LigIV and its partner protein XRCC4 (7) (Fig. 2C), there is a contradictory report that the XRCC4 NLS is required for the nuclear localization of both XRCC4 and LigIV (8) (Fig. 2C). Initiation of translation at different AUG codons within the ubiquitously expressed LigIIIα mRNA generates two LigIIIα polypeptides with different Ntermini (2) (Fig. 1A). The additional N-terminal sequence of the longer LigIIIα polypeptide forms an amphipathic α helix that engages with the mitochondrial protein import machinery, resulting in translocation into mitochondrial matrix and concomitant removal of the amphipathic α helix (Fig. 2B). Lacking both a mitochondrial targeting sequence and a NLS, the shorter LigIIIα polypeptide appears to be dependent upon forming a complex with its partner protein, XRCC1 and utilizing the XRCC1 NLS to gain entry to the nucleus (2) (Fig. 2B). In male germ cells, alternative splicing generates LigIIIβ mRNA that encodes a version of DNA ligase III, in which the C-terminal BRCT domain is replaced by a short peptide with a high proportion of positively charged amino acids that acts as an NLS (Fig. 2B) (2). The role of LigIIIβ in male germ cells has not been elucidated.
Fig. 2. Subcellular localization and functions of human DNA ligases.
A. After translation of LigI mRNA in the cytoplasm, LigI polypeptide is directed to the nucleus via an N-terminal NLS (white). B. Two different AUG codons in LigIIIα mRNA are used to initiate translation. Translation initiation at the first AUG generates a LigIIIα polypeptide with an N-terminal MLS (brown) that enables LigIIIIα but not XRCC1 to enter mitochondria. An internal AUG, which is preferentially utilized to initiate translation by the ribosome, generates a LigIIIα polypeptide lacking the MLS. This polypeptide, which also lacks a NLS, forms a complex with XRCC1 and is targeted to the nucleus via the XRCC1 NLS (white). C. Both LigIV and its partner protein XRCC4 have NLSs (white). There are contradictory reports as to whether LigIV is responsible for the nuclear localization of XRCC4 or vice versa. D. The participation of human DNA ligases in nuclear (right panel) and mitochondrial (left panel) DNA replication and repair is indicated with the major enzyme are bolded. While the original proposed mechanism of mitochondrial DNA replication does not require multiple ligation events, other mechanisms have been proposed.
As the sole DNA ligase in mitochondria (2), LigIIIα is responsible for all DNA joining events during the replication and repair of mitochondrial DNA (Fig. 2D). Analysis of the roles of the three nuclear DNA ligases has revealed significant functional overlap. While the role of LigI as the predominant DNA ligase joining Okazaki fragments during DNA replication has been well established (6), it is also now evident that LigIIIα enables cells to replicate when LigI activity is reduced or absent (9,10) (Fig. 2D). A similar functional redundancy between LigI and LigIIIα also occurs in the repair of base lesions by base excision (BER) and nucleotide excision repair (NER) as well as single strand break repair (SSBR, Fig. 2D) (6). Since the steady state levels of LigI correlate with cell proliferation and the association of LigI with the replication machinery (6), it seems reasonable to assume that the LigI subpathways of excision repair occur during the S phase of the cell cycle and are likely closely linked with DNA replication whereas the LigIIIα subpathways operate throughout the cell cycle and in non-dividing cells. While there is no direct evidence linking a specific DNA ligase with either DNA mismatch repair or recombinational repair of DNA double strand breaks (DSBs), the association of these repair pathways with DNA replication make LigI the obvious candidate to be the predominant DNA ligase involved in these pathways. In contrast to the versatility of LigI and LigIIIα in DNA replication and repair, the contributions of LigIV are limited to the repair of DSBs by non-homologous end-joining (NHEJ) (11). This pathway operates throughout the cell cycle and in non-dividing cells as well as in immune cells during the DSB-induced rearrangement of genes encoding immunoglobulins and T cell receptors (11). Finally, there are LigI- and LigIIIα-dependent alternative end joining pathways that make only a minor contribution to DSB repair under normal circumstances but are important for viability in cells deficient in either NHEJ or recombinational repair (12).
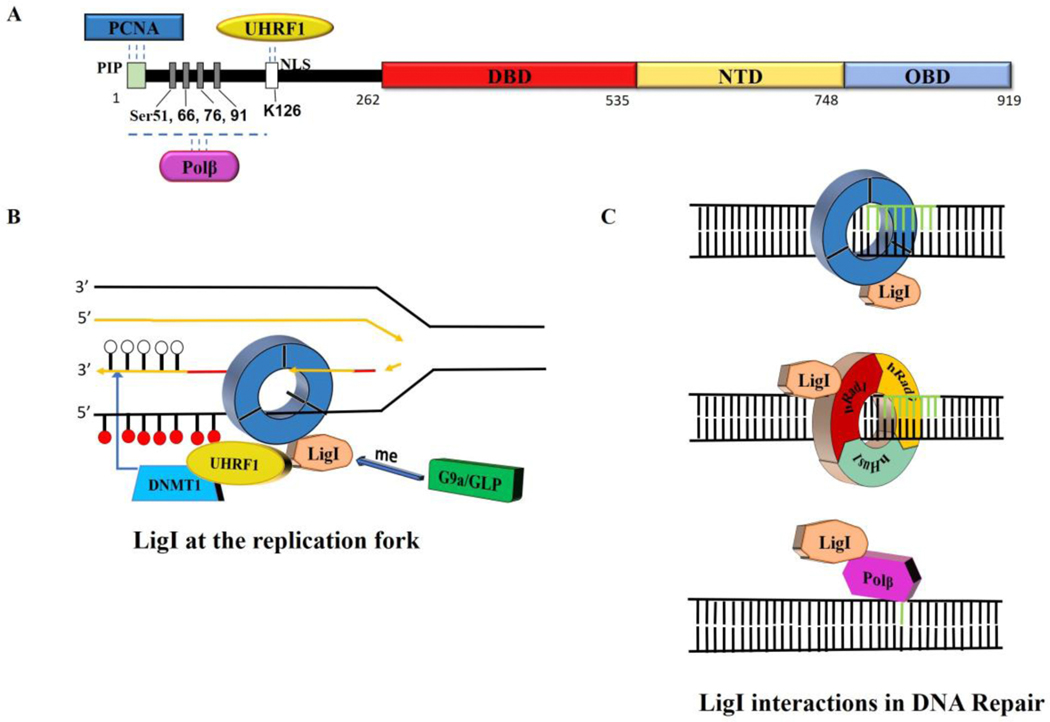
The participation of human DNA ligases in different DNA metabolic pathways is mediated by interactions with specific protein partners. For LigI (Fig. 3A), an interaction with the homotrimeric DNA sliding clamp, proliferating cell nuclear antigen (PCNA), is critical for the recruitment of LigI to replication foci as well as the joining of Okazaki fragments (Fig. 3B) and its participation in NER, long patch BER and presumably subpathways SSBR and alt-EJ (Fig. 3C) (6). LigI also interacts with the clamp loader for PCNA, replication factor C (RFC), and the checkpoint clamp Rad9-Rad1-Hus1 (Fig. 3C) and clamp loader Rad17-RFC (6). In addition, LigI interacts with Polβ within a BER complex from bovine testes (Fig. 3A and C), suggesting a role for LIgI in short patch BER (13,14). Recently, an unexpected methylation-dependent interaction between LIgI and Uhrf1 was detected and shown to play a role in recruiting the maintenance DNA methylase, Dnmt1, at the replication fork, providing an interesting mechanistic link between the joining of Okazaki fragments and DNA methylation (Fig. 3A and B) (15).
Fig. 3. LigI protein partners.
A. Linear schematic of LigI polypeptide showing the N-terminal binding sites for PCNA, Polβ and UHRF1 as well phosphorylated serine residues and an acetylated lysine residue. B. Interaction of LigI with PCNA is critical for the recruitment of LIgI to replication foci and the efficient joining of Okazaki fragments. Methylation of LigI by G9a is required for an interaction with UHRF1/DNMT1 that enables DNMT1 to methylate the newly synthesized DNA at the replication fork (white circles unmethylated cytosine; red circles, methylated cytosine. C. Upper panel, interaction of LigI with PCNA is critical for the repair of non-bulky lesions by the long patch BER pathway and the S-phase specific subpathway of NER. Middle panel, interaction of LigI with the hRad9-hRad1-hHus1 clamp stimulates DNA joining during long patch BER and may also occur in NER. Lower panel, LigI interacts with Polβ within a BER complex, suggesting a role for LigI in short patch BER.
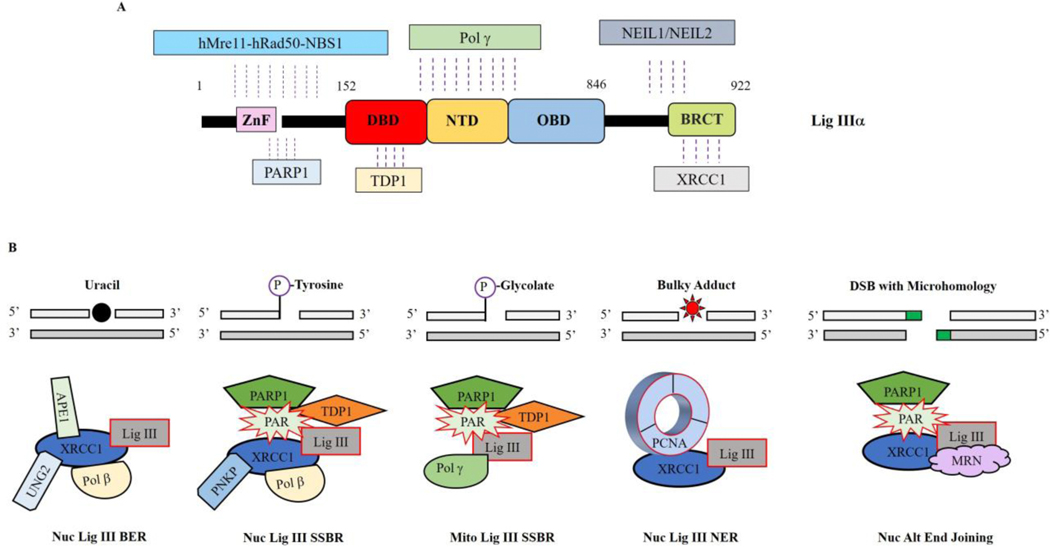
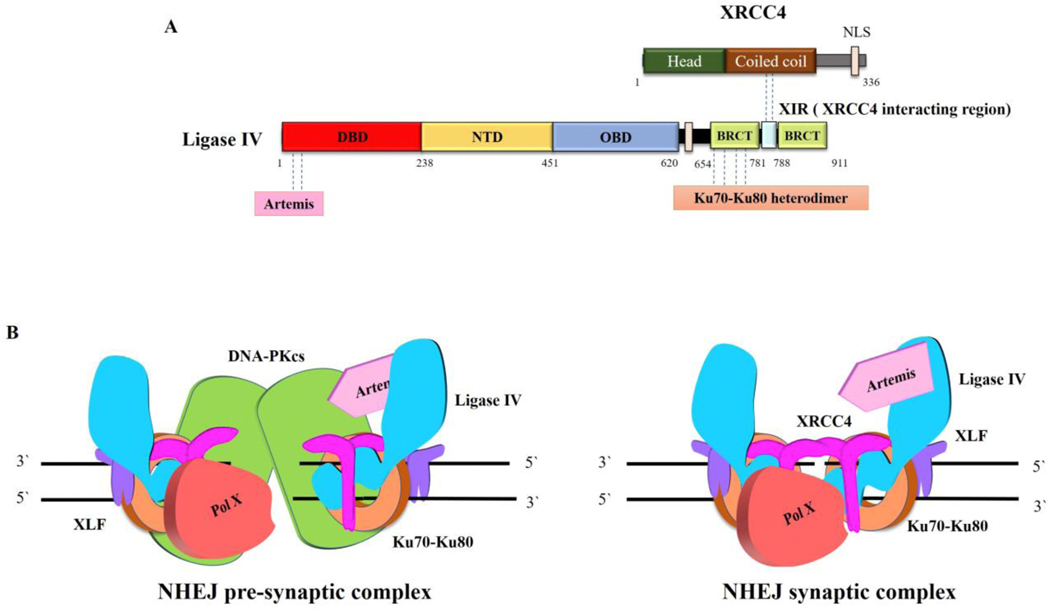
While the protein-protein interactions involving LigI are transient and most likely regulated by post translational modifications, both LigIV and nuclear LigIIIα form stable complexes with a protein partner and are unstable in the absence of their partner. As noted above, nuclear LigIIIα interacts with the DNA repair scaffold protein XRCC1 (Fig. 4A) with this heterodimer engaging with multiple other DNA repair proteins in the short patch BER, NER, SSBR and alt-EJ (Fig. 4B) (2,16). While most proteins, such as UNG2, APE1, APTX, PCNA, PNKP and Pol β, interact with XRCC1 (Fig. 4B) (16), Tdp1 interacts only with LigIIIα (Fig. 4A and B)(17), whereas MRE11-RAD50-NBS1, NEIL1/NEIL2 and PARP1 interact with both LigIIIα and XRCC1 (Fig. 4A and B) (2,16). It is likely that all these interactions contribute to nuclear DNA repair. Much less is known about how LigIIIα functions in mitochondrial DNA metabolism in the absence of XRCC1. An interaction with DNA polymerase γ (Fig. 4A) provides a specific link with mitochondrial DNA replication and repair (18) with interactions with other repair proteins, such as PARP1 and Tdp1 that appear to function in both nuclear and mitochondrial compartments, presumably contributing to mitochondrial DNA repair (Fig. 4B) (19,20). The complex formed between DNA ligase IV and an XRCC4 dimer (Fig. 5A) is recruited at an early stage of DSB repair by NHEJ, binding to the Ku heterodimer at a DNA end (Fig. 5A and B) (11). Notably, this initial recruitment appears to occur at the same as other Ku interacting proteins, DNA PKcs and XLF (11), forming a large complex that juxtaposes DNA ends through DNA PKcs-DNA PKcs interactions (Fig. 5B, left panel) (21). It is generally assumed that there is a transition involving DNA PK autophosphorylation to a synaptic complex containing Ku, LigIV/XRCC4 and XLF in which the DNA ends are aligned and accessible to LigIV (Fig. 5B, right panel) as well as end processing activities such as Artemis that binds to LigIV (Fig. 5A) and Pol X family DNA polymerases (11).
Fig. 4. LigIIIα protein partners.
A. Linear schematic of LigIIIα polypeptide showing the binding sites for hMre11-Rad50-Nbs1, PARP1, TDP1, Pol γ, NEIL1/NEIL2 and XRCC1. B. LigIIIα/XRCC1 complexes involved in nuclear short patch base excision repair (BER), short patch single strand break repair (SSBR), nucleotide excision repair subpathway operating throughout the cell cycle (NER) and alternative (ALT) end-joining. In these complexes, XRCC1 acts as a flexible scaffold that links end-processing enzymes with the LigIII catalytic region. While similar complexes lacking XRCC1, such as the predicted mitochondrial SSBR complex, are likely to be involved in mitochondrial DNA repair, DNA metabolism in this organelle is less well characterized than in the nucleus.
Fig. 5. LigIV protein partners.
A. Linear schematic of LigIV polypeptide showing the binding sites for Artemis, Ku70/Ku80 and XRCC4. B. The Ku heterodimer ring (orange) threads onto DSB ends and then recruits other core NHEJ components, DNA PKcs (green), Ligase IV (blue)/XRCC4 (magenta) and XLF (purple) to form a pre-synaptic complex in which the DNA ends are brought together but no aligned (left panel). Other factors, such as Artemis (cyan) and DNA polymerase family X members μ or λ (red), that may be needed to modify ends prior to ligation are also recruited. In the pre-synaptic complex, the DNA ends are not accessible for processing and ligation. There is predicted to be a transition involving autophosphorylation of DNA PKcs that results in a conformational change in DNA PKcs and/or the its departure from the complex, resulting in the formation of the synaptic complex (right panel), in which the DNA ends are aligned and accessible to interactions with Ligase IV and end processing enzymes.
In conclusion, the study of human DNA ligases has provided insights into DNA replication, DNA methylation and DNA repair pathways. While the interaction of the DNA ligase catalytic region with nicked DNA is well characterized and many of the pathways have been reconstituted with purified proteins, more work is needed to understand how the highly flexible regions flanking the catalytic regions, which are reversibly modified by posttranslational modification, contribute to dynamic replication and repair complexes. In addition, there is limited information about the contributions of the DNA ligases in different cell types, particularly terminally differentiated cells. For example, while the immunodeficiency in Lig4 syndrome is due to a defect in V(D)J recombination, the molecular defect underlying the immunodeficiency in Lig1 syndrome is not obvious (5). Finally, there is growing evidence that that DNA ligase expression is frequently altered in cancer and that the changes in expression are indicative of altered DNA repair (5). This, together with promising results showing that DNA ligase inhibitors can be used to preferentially target cancer cell lines, suggest that DNA ligase inhibitors may have utility as cancer therapeutics in precision medicine (5, 22).
Acknowledgements
We apologize to colleagues whose primary publications have not been cited because of limitations in the number of references. Work in the Tomkinson laboratory was supported by the University of New Mexico Comprehensive Cancer Center and National Institute of Health Grants R01 GM57479 (to A.E.T.), R01 ES012512 (to A.E.T.), R01 GM57251 (to A.E.T.) P01 CA92584 and P30 CA118100.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ellenberger T. and Tomkinson AE (2008) Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem, 77, 313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tomkinson AE and Sallmyr A. (2013) Structure and function of the DNA ligases encoded by the mammalian LIG3 gene. Gene, 531, 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ho CK, Wang LK, Lima CD and Shuman S. (2004) Structure and mechanism of RNA ligase. Structure (Camb), 12, 327–339. [DOI] [PubMed] [Google Scholar]
- [4].Shuman S. and Schwer B. (1995) RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol Microbiol, 17, 405–410. [DOI] [PubMed] [Google Scholar]
- [5].Tomkinson AE, Naila T. and Khattri Bhandari S. (2020) Altered DNA ligase activity in human disease. Mutagenesis, 35, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Howes TR and Tomkinson AE (2012) DNA ligase I, the replicative DNA ligase. Subcell Biochem, 62, 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Francis DB, Kozlov M, Chavez J, Chu J, Malu S, Hanna M. and Cortes P. (2014) DNA Ligase IV regulates XRCC4 nuclear localization. DNA Repair (Amst), 21, 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fukuchi M, Wanotayan R, Liu S, Imamichi S, Sharma MK and Matsumoto Y. (2015) Lysine 271 but not lysine 210 of XRCC4 is required for the nuclear localization of XRCC4 and DNA ligase IV. Biochem Biophys Res Commun, 461, 687–694. [DOI] [PubMed] [Google Scholar]
- [9].Han L, Masani S, Hsieh CL and Yu K. (2014) DNA ligase I is not essential for Mammalian cell viability. Cell Rep, 7, 316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Le Chalony C, Hoffschir F, Gauthier LR, Gross J, Biard DS, Boussin FD and Pennaneach V. (2012) Partial complementation of a DNA ligase I deficiency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells. Cell Mol Life Sci, 69, 2933–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pannunzio NR, Watanabe G. and Lieber MR (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem, 293, 10512–10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sallmyr A. and Tomkinson AE (2018) Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J Biol Chem, 293, 10536–10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dimitriadis EK, Prasad R, Vaske MK, Chen L, Tomkinson AE, Lewis MS and Wilson SH (1998) Thermodynamics of human DNA ligase I trimerization and association with DNA polymerase beta. J Biol Chem, 273, 20540–20550. [DOI] [PubMed] [Google Scholar]
- [14].Prasad R., Singhal RK., Srivastava DK., Molina JT., Tomkinson AE. and Wilson SH. (1996) Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J Biol Chem, 271, 16000–16007. [DOI] [PubMed] [Google Scholar]
- [15].Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S, Kirsh O, Amouroux R, Dohmae N, Suzuki T. et al. (2017) Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to Replicating DNA and Regulates DNA Methylation. Mol Cell, 67, 550–565 e555. [DOI] [PubMed] [Google Scholar]
- [16].Caldecott KW (2019) XRCC1 protein; Form and function. DNA Repair (Amst), 81, 102664. [DOI] [PubMed] [Google Scholar]
- [17].El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR and Caldecott KW (2005) Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature, 434, 108–113. [DOI] [PubMed] [Google Scholar]
- [18].De A. and Campbell C. (2007) A novel interaction between DNA ligase III and DNA polymerase gamma plays an essential role in mitochondrial DNA stability. Biochem J, 402, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Das BB, Dexheimer TS, Maddali K. and Pommier Y. (2010) Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci U S A, 107, 19790–19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Szczesny B., Brunyanszki A., Olah G., Mitra S. and Szabo C. (2014) Opposing roles of mitochondrial and nuclear PARP1 in the regulation of mitochondrial and nuclear DNA integrity: implications for the regulation of mitochondrial function. Nucleic Acids Res, 42, 13161–13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].DeFazio LG, Stansel RM, Griffith JD and Chu G. (2002) Synapsis of DNA ends by DNA-dependent protein kinase. Embo J, 21, 3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tomkinson AE, Howes RL and Wiest NE (2013) DNA ligases as therapeutic targets Transl Cancer Res, 2, 1219. [PMC free article] [PubMed] [Google Scholar]
- [23].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]