Abstract
BACKGROUND
In degenerative intervertebral disc (IVD), an unfavorable IVD environment leads to increased senescence of nucleus pulposus (NP)-derived mesenchymal stem cells (NPMSCs) and the inability to complete the differentiation from NPMSCs to NP cells, leading to further aggravation of IVD degeneration (IDD). Urolithin A (UA) has been proven to have obvious effects in delaying cell senescence and resisting oxidative stress.
AIM
To explore whether UA can alleviate NPMSCs senescence and to elucidate the underlying mechanism.
METHODS
In vitro, we harvested NPMSCs from rat tails, and divided NPMSCs into four groups: the control group, H2O2 group, H2O2 + UA group, and H2O2 + UA + SR-18292 group. Senescence-associated β-Galactosidase (SA-β-Gal) activity, cell cycle, cell proliferation ability, and the expression of senescence-related and silent information regulator of transcription 1/PPAR gamma coactivator-1α (SIRT1/ PGC-1α) pathway-related proteins and mRNA were used to evaluate the protective effects of UA. In vivo, an animal model of IDD was constructed, and X-rays, magnetic resonance imaging, and histological analysis were used to assess whether UA could alleviate IDD in vivo.
RESULTS
We found that H2O2 can cause NPMSCs senescence changes, such as cell cycle arrest, reduced cell proliferation ability, increased SA-β-Gal activity, and increased expression of senescence-related proteins and mRNA. After UA pretreatment, the abovementioned senescence indicators were significantly alleviated. To further demonstrate the mechanism of UA, we evaluated the mitochondrial membrane potential and the SIRT1/PGC-1α pathway that regulates mitochondrial function. UA protected mitochondrial function and delayed NPMSCs senescence by activating the SIRT1/PGC-1α pathway. In vivo, we found that UA treatment alleviated an animal model of IDD by assessing the disc height index, Pfirrmann grade and the histological score.
CONCLUSION
In summary, UA could activate the SIRT1/PGC-1α signaling pathway to protect mitochondrial function and alleviate cell senescence and IDD in vivo and vitro.
Keywords: Urolithin A, Mitochondrial function, Oxidative stress, Senescence, Nucleus pulposus-derived Mesenchymal stem cells, The silent information regulator of transcription 1/PPAR gamma coactivator-1α pathway
Core Tip: In degenerative intervertebral disc (IVD), an unfavorable IVD environment leads to increased senescence of nucleus pulposus-derived mesenchymal stem cells (NPMSCs), which seriously affects endogenous repair of IVD. Urolithin A (UA) alleviated oxidative stress-induced NPMSCs senescence by activating the silent information regulator of transcription 1/PPAR gamma coactivator-1α signaling pathway and protecting mitochondrial function in vitro. UA could also delay extracellular matrix degradation and IVD degeneration (IDD) in vivo. The results provide the possibility to promote endogenous repair and retard IDD.
INTRODUCTION
Low back pain seriously affects the quality of life and increases the economic burden on families and society[1,2]. Intervertebral disc degeneration (IDD) is the main pathogenic factor of low back pain, but its pathological mechanism has not yet been elucidated[3]. Therefore, exploring the pathological mechanism of IDD and seeking new methods for the prevention and treatment of degenerative disc diseases is of great significance to human health and social development.
Nucleus pulposus (NP) cells play an important role by secreting a large amount of extracellular matrix (ECM) such as aggrecan and collagen type II to maintain the normal physiological function of the intervertebral disc (IVD) under physiological conditions. However, NP cells are terminal cells with low proliferation ability and no differentiation and self-renewal ability. In degenerative IVD, the number of NP cells is reduced, and their function is impaired, leading to a decrease in ECM secretion and aggravation of IDD. Endogenous repair is a specific repair mediated by tissue-specific stem cells and has been found to exist in a variety of tissues, such as the skin, liver and nervous system[4,5]. In 2007, Risbud et al[6] isolated and identified NP-derived mesenchymal stem cells (NPMSCs) in degenerative IVD, which provided a basis for the endogenous repair of IDD. However, the unfavorable microenvironment of degenerative IVD, such as inflammation, oxidative stress, and increased catabolism, leads to increased senescence and apoptosis of NPMSCs, which seriously affects endogenous repair[7]. Therefore, rescuing the activity of NPMSCs and delaying cell senescence is of great significance to alleviate IDD.
To date, natural metabolically active products have been widely found as important sources for drug discovery in antiaging and senescence-related diseases. Urolithin is a type of dibenzopyran-6-one derivative with different phenolic hydroxyl groups produced by intestinal microbial metabolism from foods rich in ellagitannins (pomegranate, strawberry, walnut, raspberry, etc.)[8]. Among them, urolithin A (UA) (Figure 1) was the first to be isolated and identified from the feces and urine of mice fed ellagic acid[9]. Previous studies found that UA can show biological effects, such as regulating estrogen secretion and antioxidant and anti-inflammatory activities[10-12]. Recently, the antiaging effect of UA has drawn considerable attention. Ryu et al[13] found that UA has a unique effect in inducing mitophagy, prolonging the lifespan of C. elegans and increasing muscle function in rodents. A previous study also found that UA could exert antiapoptotic and antiaging effects on NP cells[14,15].
Figure 1.

Chemical structure of Urolithin A.
However, there are few studies of the protective effect of UA on NPMSCs to date. In this study, we investigated whether UA could alleviate H2O2-induced NPMSCs senescence in vitro and in IDD animal models in vivo and elucidated the mechanisms involved in this progress.
MATERIALS AND METHODS
Isolation and culture of NPMSCs
This study was approved by the Ethical Committee of the Clinical Medical College of Yangzhou University (SBYY2020-023). Sprague-Dawley (SD) rats (weight, 200-300 g; age, 4-6 mo) were purchased from the Shanghai Institute of Family Planning Science [License No. SCXK (Hu) 2018-0006]. NPMSCs were harvested from the coccygeal IVD tissues of SD rats. Then, the NP tissues were isolated under a dissecting microscope and digested in 0.2% type II collagenase (Gibco, United States, catalog No. 17101015) for 12 h at 37 °C with 5% CO2. Then, the obtained cells and partially digested tissues were washed with phosphate-buffered saline (PBS) twice, and centrifuged at 1000 r/min for 5 min, and then cultured in Mesenchymal Stem Cell Complete Medium (Cyagen, United States, catalog No. RASMX-90011) at 37 °C with 5% CO2. The culture medium was changed every three days. The cells were passaged at a 1:3 ratio at 80%-90% confluence. NPMSCs used in the followed study was passage 3.
Surface marker identification of NPMSCs
The mesenchymal stem cell (MSC)-associated surface markers were examined by immunofluorescent staining. Cell slides with a diameter of 25 mm containing polylysine were placed in a 12-well plate, and NPMSCs were seeded and cultured in MSC complete medium. Then, the cells were fixed with 4% paraformaldehyde for 15 min and washed twice with PBS containing 0.5% Triton X-100 for 15 min. Then, the cells were blocked with 10% bovine serum albumin for 1 h at 37 °C and incubated with primary antibodies against CD 105 (Proteintech, China, catalog No. 10862-1-AP), CD90 (ABclonal, China, catalog No. A2126), CD73 (ABclonal, China, catalog No. A2029), CD34 (ABclonal, China, catalog No. A7429) and CD45 (ABclonal, China, catalog No. A2115) (1:100) at 4 °C overnight. The cell slides were washed twice with PBS and then incubated with secondary antibodies (Abcam, United Kingdom, catalog No. ab150077, ab150078) (1:500) for 1 h at room temperature. After treatment with the antifade mounting medium with 4’,6-diamidino-2’-phenylindole for 10 min, the cell slides were observed and recorded using a fluorescence microscope (Leica, Wetzlar, Germany).
Multilineage differentiation
To demonstrate the multilineage differentiation potential of NPMSCs, osteogenic, adipogenic and chondrogenic differentiation was induced. NPMSCs were seeded in 6-well plates and cultured until reaching approximately 80% confluency. Then, the culture medium was changed to osteogenic, cartilage and adipogenic differentiation medium (Cyagen, China, catalog No. RASMX-90021, RASMX-9004, RASMX-90031), and the medium was changed every 3 d according to the manufacturer’s instructions. After reaching the deadline of induction, the cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min, and then the cells were processed with oil red O, alizarin red and alcian blue. Finally, the staining results were observed and imaged under a fluorescence microscope.
Cell viability assay
NPMSCs were seeded in 96-well plates at a density of 2 × 103 cells per well and incubated in complete medium overnight at 37 °C with 5% CO2. Then, NPMSCs were treated with different concentrations ((0-80 μM, 0-48 h) of UA (MedChem Express, China, catalog No. 1143-70-0). After that, 10% cell counting kit-8 (CCK-8) (Dojindo, Japan, catalog No. CK04) was added to each well at different time points, and the optical density (OD) value was read after 1 h of incubation at 480 nm by a microplate reader (Bio-Rad, United States). Cell viability was calculated as follows: Cell viability (of control) = [(Ae-Ab)/(Ac-Ab)]. Ae, Ab, and Ac represent the OD value of the treatment, blank and control groups, respectively. Similarly, to further determine whether UA works through the silent information regulator of transcription 1/PPAR gamma coactivator-1α (SIRT1/PGC-1α) pathway, different concentrations (0-160 μM, 0-36 h) of the PGC-1α inhibitor SR-18292 (MedChem Express, China, catalog No. HY-101491) were cocultured with NPMSCs.
NPMSCs were divided into four groups in the following examinations: (1) Control group; (2) H2O2 group (80 μM H2O2); (3) H2O2 + UA group (80 μM H2O2 + 20 μM UA); and (4) H2O2 + UA + SR-18292 group (80 μM H2O2 + 20 μM UA + 20 μM SR-18292).
Cell proliferation assay
NPMSCs (5 × 104 cells/well) were seeded in a 12-well plate and cultured in MSC complete medium. An EdU Cell Proliferation Kit (Beyotime, China, catalog No. C0071S) was used to detect cell proliferation. Subsequently, NPMSCs were incubated with EdU for 2 h and fixed with 4% paraformaldehyde for 15 min, and then cells were incubated with 0.3% Triton X-100 for 10 min according to the manufacturer’s instructions. Then, the cells were incubated with Click Reaction Mixture for 30 min and then incubated with Hoechst 33342 for 10 min in the dark. Finally, cells were observed and recorded using a fluorescence microscope and analyzed by ImageJ software (NIH, United States).
Cytotoxicity assay
Cytotoxicity was measured by the lactate dehydrogenase (LDH) activity in the supernatant using the LDH cytotoxicity assay kit (Beyotime, China, catalog No. C0016) according to the protocol. NPMSCs were seeded in 96-well plates (5 × 103 cells/well) and incubated in complete medium overnight at 37 °C with 5% CO2. Then, NPMSCs were treated with 80 μM H2O2, 80 μM H2O2 + 20 μM UA and 80 μM H2O2 + 20 μM UA + 20 μM SR-18292 in the H2O2 group, H2O2 + UA group and H2O2 + UA + SR-18292 group, respectively. After that, 10% LDH release reagent was added to each well and incubated for 1 h. Then, the supernatant was transferred to a new 96-well plate and mixed with LDH detection working solution in the dark for 30 min. Finally, the OD value was detected at 490 nm by a microplate reader. The cytotoxicity was calculated as follows: Cytotoxicity (of control) = [(Ae-Ab)/(Ac-Ab)]. Ae, Ab, and Ac represent the OD values of the treatment, blank and control groups, respectively.
Senescence-associated β-Galactosidase staining
NPMSCs were seeded in a 6-well plate (1 × 104 cells/well), and senescence-associated β-Galactosidase (SA-β-Gal) staining was performed according to the manufacturer’s instructions from the SA-β-Gal staining Kit (Beyotime, China, catalog No. C0602). NPMSCs were observed under a fluorescence microscope and analyzed by ImageJ software.
Cell cycle assay
NPMSCs were seeded in 6-well plates with serum-free medium overnight. After that, cells were collected and fixed with 75% ethanol overnight. Then, the cells were incubated with a mixed solution of propidium iodide (PI) dye and RNase A (Keygen, China, catalog No. KGA511) for 30 min, and the cell cycle phases were analyzed by flow cytometry (BD Company, United States).
JC-1 assay for mitochondrial membrane potential
Mitochondrial membrane potential (MMP) was measured using the JC-1 (5,5’,6,6’-tetrachloro-1,1’,3,3’-tetraethylbenzimidazolcarbocyanine iodide) Detection Kit (Keygen, China, catalog No. KGA603). NPMSCs from different groups were washed with PBS and incubated with 2 μM JC-1 dye for 20 min. Then, the cells were washed twice with incubation buffer and observed using a fluorescence microscope. The ratio of green (depolarization) to red (polarized) fluorescence intensity was calculated using ImageJ.
Reactive oxygen species
The level of intracellular reactive oxygen species (ROS) in NPMSCs was measured by a ROS detection fluorescent probe-DHE kit (Keygen, China, catalog No. KGAF019). After different interventions in a 12-well plate, NPMSCs were washed twice with PBS and incubated with 20 μM DHE for 1 h at 37 °C according to the manufacturer’s instructions. Then, the cells were observed using a fluorescence microscope and analyzed by ImageJ.
Quantitative real-time polymerase chain reaction
Total RNA was extracted using TRIzol reagent (Invitrogen, United States, catalog No. 15596-026). Reverse transcription from whole RNA to complementary DNA (cDNA) and amplification of the cDNA were performed using a Prime Script-RT reagent kit (Vazyme Biotech, China, catalog No. R123-01) and SYBR Premix Ex Taq (Vazyme Biotech, China, catalog No. Q111-02) according to the manufacturer’s instructions. The expression of target genes of NPMSCs in different groups was calculated by the comparative Ct method. The primers were designed according to the sequences in GenBank using Prime 5.0 software and are listed in Table 1.
Table 1.
Sequences of primers used for real-time PCR
|
Gene
|
Primer sequence
|
| GAPDH | Forward 5'-CTGGAGAAACCTGCCAAGTATG-3'Reverse 5'-GGTGGAAGAATGGGAGTTGCT-3' |
| P16 | Forward 5'-CCGATACAGGTGATGATGATGG-3'Reverse 5'-CGGAGGAGAGTAGATACCGCAAA-3' |
| P21 | Forward 5'-AGTTGGAGCTGGTGGCGTAG-3'Reverse 5'-AATACACAAAGAAAGCCCTCCC-3' |
| SIRT1 | Forward 5'-AGATTTCAAGGCTGTTGGTTCC-3'Reverse 5'-CAGCATCATCTTCCAAGCCATT-3' |
| PGC-1α | Forward 5'-GAGAAGCGGGAGTCTGAAAGG-3'Reverse 5'-GTCACAGGTGTAACGGTAGGTAATG-3' |
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; SIRT1: Silent information regulator of transcription 1; PGC-1α: PPAR gamma coactivator-1α.
Western blot assay
Total protein was extracted from NPMSCs by Whole Cell Lysis Assay (Keygen, China, catalog No. KGP250), and the protein concentration was measured using the BCA protein assay kit (Beyotime, China, catalog No. P0010). Then, an equal protein sample of each group was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane. After that, the membranes were blocked with 5% nonfat milk for 2 h at room temperature and then incubated with primary antibodies against β-actin (Proteintech, China, catalog No. 20536-1-AP) (1:3000), p16 (Proteintech, China, catalog No. 10883-1-AP) (1:1000) and p21 (Proteintech, China, catalog 10355-1-AP) (1:1000) overnight at 4 °C. After washing three times with Tris-buffered saline and 0.1% Tween 20 (TBST), the membranes were incubated with secondary antibodies (Proteintech, China, catalog No. SA00001-2) (1:5000) for 2 h on a shaker at room temperature. Then, the membranes were visualized using an enhanced chemiluminescence system, and the relative amount of protein was analyzed using ImageJ software.
IDD animal model induction
Fifteen SD rats (weight, 200-300 g; age, 4-6 mo) were randomly divided into three groups (n = 5 per group): the control group (no operation), IDD group (punctured and DMSO treatment), and UA group (punctured and UA treatment). The SD rat IDD model was established according to a previous method[16]. Briefly, the rats were anesthetized by an overdose of pentobarbital. After sterilization with povidone iodine, the coccygeal IVD (CO6-7) was percutaneously punctured by a 21 G needle at a depth of 5 mm, followed by rotation at 360° and holding for 30 s. The UA group was given water containing UA (25 mg/kg/d, dissolved in DMSO and diluted in water) for 4 wk from the first day after surgery[13]. The control group and IDD group were given an equivalent volume of DMSO for 4 wk.
Radiographic and magnetic resonance imaging evaluation
Radiographic and MRI scans were taken prepuncture and 4 wk after puncture, respectively. The rats were placed in a prone position after anesthesia by inhalation of 2% isoflurane. X-ray scans were performed, and the disc height index (DHI) was measured by ImageJ software[17].
The signal and structural change of the IVD were obtained by a 3.0-T clinical MR scanning system (Philips Intera Achieva 3.0 MR, Netherlands). Briefly, SD rats were maintained after inhalation of 2% isoflurane and then placed in a prone position. Sagittal T2-weighted images were evaluated according to the Pfirrmann grade[18].
Histologic analysis
All SD rats were euthanized by an overdose of pentobarbital after 4 wk of puncture. The IVD specimens were harvested and fixed with 4% paraformaldehyde, decalcified with 10% ethylenediaminetetraacetic acid solution, and embedded in paraffin. The specimens were cut into 5-μm sections, and the slices were stained with hematoxylin-eosin (HE), toluidine blue and safranin-O stains. Histologic images of HE were evaluated following histologic grading scale criteria reported by Norcross et al[19].
Immunofluorescent staining
After the rats were killed, IVD specimens were harvested and cut into 5 μm sections with a freezing microtome (Leica, Wetzlar, Germany). The sections were then fixed in 4% paraformaldehyde for 15 min and washed twice with PBS. Then, the sections were blocked with 10% bovine serum albumin for 1 h at 37 °C and incubated with primary antibodies at 4 °C overnight: rabbit polyclonal anti-collagen type II (ABclonal, China, Catalog No. A1560) (1:1000) and anti-aggrecan (ABclonal, China, Catalog No. A8536) (1:1000). After that, the sections were washed twice with PBS and incubated with secondary antibodies (Abcam, United Kingdom, catalog No. ab150077, ab150078) (1:500) for 1 h at room temperature in the dark. The sections were photographed by a fluorescence microscope and analyzed by ImageJ software.
Statistical analysis
All data were analyzed by Statistical Package for the Social Sciences (SPSS) software (version 26; IBM, Chicago, Illinois). The quantitative data are expressed as the mean ± SD. The data of multiple independent groups were analyzed by one-way ANOVA. Student’s t-test was used to analyze the differences between the two groups. P value < 0.05 was significant.
RESULTS
Identification of NPMSCs
The cells isolated from the rat coccygeal IVD presented with a long spindle shape and grew in flower formation (Figure 2A). As shown in Figure 2B, MSC-associated surface markers were identified by cellular immunofluorescence. CD105, CD90, and CD73 showed high fluorescent expression, but CD34 and CD45 showed low fluorescent expression. The multilineage differentiation ability was confirmed by multilineage differentiation in vitro (Figure 2C). The results indicated that the cells isolated from the rat NP meet the appraisal standards of stem cells proposed by the International Society for Cellular Therapy (ISCT).
Figure 2.
Identification of nucleus pulposus-derived mesenchymal stem cells. A: Isolated primary cells presented with a long spindle shape and grew in flower formation; B: Nucleus pulposus-derived mesenchymal stem cells (NPMSCs) exhibited high fluorescent expression of CD73, CD90, and CD105, but low fluorescent expression of CD34 and CD45; C: NPMSCs was positive for Alizarin red, Oil Red O, and Alcian blue staining after induced differentiation. Scale bar = 50 μm. NPMSCs: Nucleus pulposus-derived mesenchymal stem cells.
Measurement of cell viability
An appropriate concentration of H2O2 to induce oxidative stress damage in NPMSCs was confirmed in our previous study[20]. The viability effects of UA and the PGC-1α inhibitor SR-18292 on NPMSCs were analyzed using a CCK-8 assay. NPMSCs were cocultured with different concentrations of UA and SR-18292 supplemented culture media at different time points. As shown in Figure 3A, UA (0-20 μM) showed an appropriate inhibitory effect on cell viability for 24 h, but UA (> 40 μM) exerted a significant inhibitory effect (P < 0.05). Therefore, 20 μM UA was used as the final drug intervention concentration. Similarly, after incubation with different concentrations of SR-18292 at different time points, 20 μM SR-18292 for 24 h exhibited appropriate inhibition of cell viability and was used as the final drug concentration and intervention time (Figure 3B) (P < 0.05).
Figure 3.
Cell viability assay, cell proliferation assay and cytotoxicity assay. A: Cell counting kit-8 (CCK-8) results of nucleus pulposus-derived mesenchymal stem cells (NPMSCs) treated with different concentrations and times of urolithin A (UA); B: CCK-8 results of NPMSCs treated with different concentrations and times of SR-18292; C: EdU assay results of NPMSCs in different groups. Green fluorescence represents cells in a proliferating state, and blue fluorescence represents cell nucleus (scale bar = 25 μm); D: Quantitative analysis of EdU results; E: Cytotoxicity results of NPMSCs treated with H2O2, H2O2 + UA and H2O2 + UA + SR-18292. All data are expressed as the mean ± SD. aP < 0.05, bP < 0.01 compared with control group; cP < 0.05, dP < 0.01 compared with H2O2 group; eP < 0.05, fP < 0.01 compared with H2O2 + UA group. CCK-8: Cell counting kit-8; NPMSCs: Nucleus pulposus-derived mesenchymal stem cells; UA: Urolithin A.
Measurement of cell proliferation and cytotoxicity
To elucidate the effect of H2O2, UA and SR-18292 on the proliferation ability of NPMSCs, we performed EdU staining of NPMSCs. As shown in Figure 3C-D, the EdU-positive rate of the H2O2 group was significantly lower than that of the control group (32.1% ± 5.4% vs 47.9% ± 5.8%, P < 0.05), and EdU-positive rate was increased by 12% after UA pretreatment. However, the protective effects of UA were reversed by SR-18292 (44.9% ± 5.7% vs 34.4% ± 5.9%, P < 0.05).
LDH release is regarded as an important indicator of cell membrane integrity and is widely used to assess cytotoxicity. As a common oxidative stress-inducing agent, 80 μM H2O2 still had cytotoxicity compared to the control group (P < 0.01). However, the cytotoxicity induced by H2O2 was alleviated after pretreatment with 20 μM UA, which indicates that LDH release was alleviated after UA pretreatment (P < 0.05). After treatment with SR-18292, the protective effect of UA was partly blocked (Figure 3E) (P < 0.01).
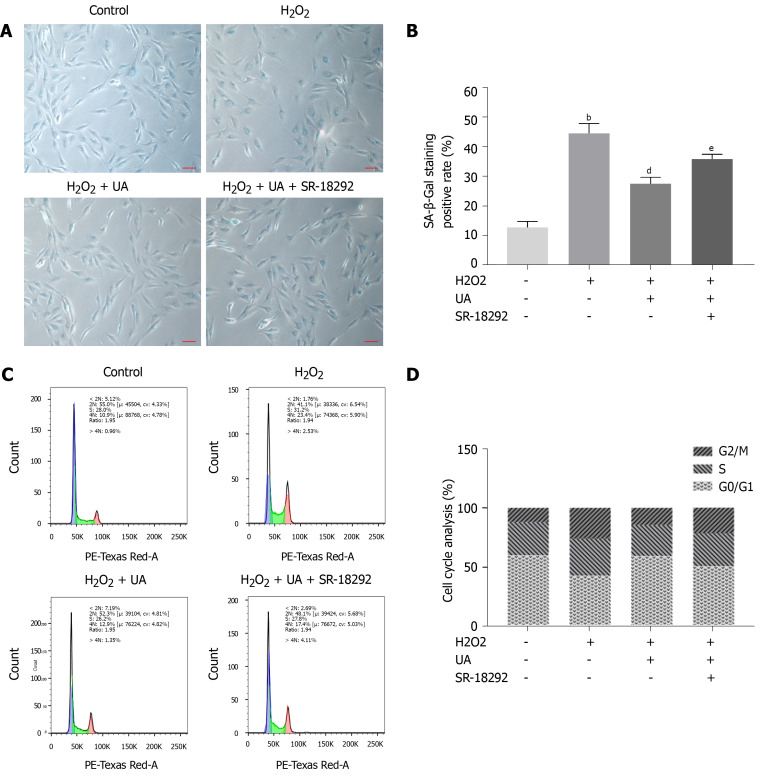
Measurement of SA-β-Gal staining
SA-β-Gal is a parameter evaluating cellular senescence, and senescence cells with high SA-β-Gal activity are stained blue. NPMSCs of the H2O2 group demonstrated a higher percentage of SA-β-Gal staining positive than the control group (P < 0.01). The percentage of positive cells was decreased after UA pretreatment (P < 0.01). The percentage of SA-β-Gal-positive cells was increased when NPMSCs were cocultured with UA and SR-18292 before H2O2 treatment (Figure 4A-B) (P < 0.05).
Figure 4.
Senescence-associated β-Galactosidase staining assay and cell cycle assay. A: Senescence-associated β-Galactosidase (SA-β-Gal) staining results of nucleus pulposus-derived mesenchymal stem cells (NPMSCs) in different groups. Senescent cells exhibit blue-stained high expression of SA-β-Gal. (scale bar = 25 μm); B: Quantitative analysis of SA-β-Gal staining results; C: Cell cycle results of NPMSCs in different groups; D: Quantitative analysis of cell cycle results. All data are expressed as the mean ± SD. aP < 0.05, bP < 0.01 compared with control group; cP < 0.05, dP < 0.01 compared with H2O2 group; eP < 0.05, fP < 0.01 compared with H2O2 + UA group. SA-β-Gal: Senescence-associated β-Galactosidase; NPMSCs: Nucleus pulposus-derived mesenchymal stem cells; UA: Urolithin A.
Measurement of cell cycle
Cell cycle arrest is one of the common features of senescent cells. As shown in Figure 4C-D, a higher percentage of NPMSCs showed cell cycle arrest in G2/M phase in the H2O2 group than in the control group. The percentage of NPMSCs arrested in G2/M phase decreased after pretreatment with UA, indicating that UA could attenuate H2O2-induced cell cycle arrest. However, the percentage of cells arrested in G2/M phase increased after treatment with SR-18292 compared with UA pretreatment alone.
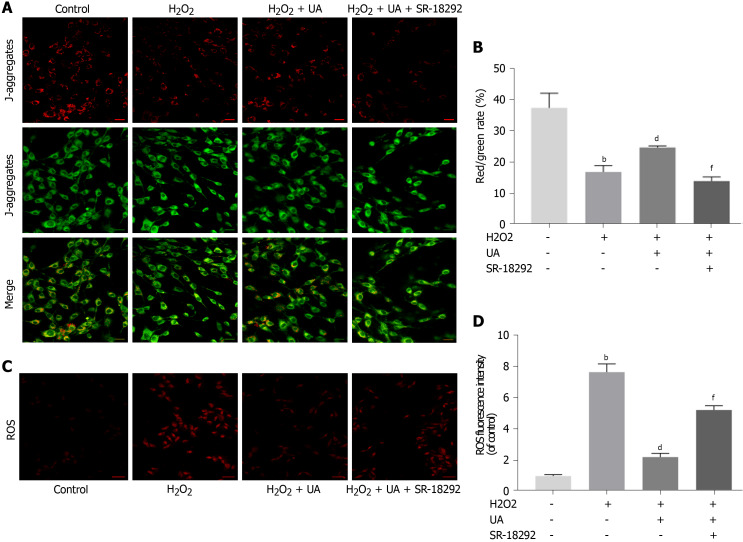
Measurement of MMP and ROS levels
The polarized MMP was stained orange-red fluorescence in the control group, whereas the red fluorescence intensity was weakened, and the green fluorescence was enhanced after H2O2 treatment (P < 0.01). Compared with the H2O2 group, the MMP of NPMSCs pretreated with UA was still in the orange-red polarization state (Figure 5A-B) (P < 0.01).
Figure 5.
Mitochondrial membrane potential assay and reactive oxygen species assay. A: Results of mitochondrial membrane potential (MMP) in different groups detected by fluorescence. Red fluorescence represents the mitochondrial aggregate JC-1 and green fluorescence indicates the monomeric JC-1 (scale bar = 50 μm); B: Quantitative analysis of MMP results; C: Results of reactive oxygen species (ROS) in different groups detected by fluorescence. Red fluorescence represents high level of ROS (scale bar = 50 μm); D: Quantitative analysis of ROS results. All data are expressed as the mean ± SD. aP < 0.05, bP < 0.01 compared with control group; cP < 0.05, dP < 0.01 compared with H2O2 group; eP < 0.05, fP < 0.01 compared with H2O2 + UA group. MMP: Mitochondrial membrane potential; ROS: Reactive oxygen species; UA: Urolithin A.
Excessive generation of ROS damages mitochondrial dynamics. As shown in Figure 5C-D, the ROS level of NPMSCs in the H2O2 group was significantly higher than that of the control group, and the ROS level of the H2O2 + UA group was significantly lower than that of the H2O2 group (P < 0.01). However, the protective effect of UA decreased after treatment with SR-18292 (P < 0.01). This result indicated that UA protected the dynamics of mitochondria through the PGC-1α signaling pathway and avoided the accumulation of ROS.
Measurement of senescence-related and SIRT1/PGC-1α pathway-related mRNA and proteins
We further evaluated the expression of senescence-related mRNA and proteins (P16 and P21) by western blotting and quantitative real-time polymerase chain reaction. As shown in Figure 6A-E, the expression of P16 and P21 in the H2O2 group was significantly increased compared with that in the control group (P < 0.05). However, the increased expression of P16 and P21 was alleviated by pretreatment with UA (P < 0.05). Moreover, pretreatment with SR-18292 weakened this protective effect of UA and decreased the expression of P16 and P21 (P < 0.05).
Figure 6.
Senescence-related and SIRT1/PGC-1α pathway-related mRNA and proteins assay. A: The expression of senescence-related proteins (P16 and P21) in different groups; B-C: Quantitative analysis of P16 and P21 protein expression results; D-E: Quantitative analysis of P16 and P21 mRNA expression results; F-G: Quantitative analysis SIRT1/PGC-1α pathway-related mRNA. All data are expressed as the mean ± SD. aP < 0.05, bP < 0.01 compared with control group; cP < 0.05, dP < 0.01 compared with H2O2 group; eP < 0.05, fP < 0.01 compared with H2O2 + UA group. SIRT1/PGC-1α: Silent information regulator of transcription 1/PPAR gamma coactivator-1α; UA: Urolithin A.
To investigate whether UA plays a role by activating the SIRT1/PGC-1α signaling pathway, related mRNA expression was evaluated. The results showed that the mRNA expression of SIRT1 and PGC-1α decreased after H2O2 treatment, but their expression was upregulated after UA treatment (P < 0.05). Then, pathway-related mRNA expression was reversed by treatment with the inhibitor SR-18292, which indicated that UA might exert a protective effect by activating the SIRT1/PGC-1α pathway (Figure 6F-G) (P < 0.05).
Radiographic and MRI evaluation
X-ray images were performed to evaluate disc height at 0 wk and 4 wk after needle puncture, and DHI was used to assess the disc height. As shown in Figure 7A-B, there was no significant difference in DHI among the three groups at 0 wk (P > 0.05). However, the DHI of the IDD group (0.040 ± 0.001) was significantly decreased compared with that of the control group (0.104 ± 0.005) at 4 wk (P < 0.01). Furthermore, the DHI of the UA group (0.068 ± 0.003) was significantly higher than that of the IDD group (P < 0.01).
Figure 7.
X-ray and magnetic resonance imaging evaluation in intervertebral disc degeneration animal models. A: The X-ray in different groups at 0 wk and 4 wk after puncturing; B: Quantitative analysis of the disc height index in different groups; C: The magnetic resonance imaging results of different groups at 0 wk and 4 wk after puncture; D: Quantitative analysis of Pfirrmann grades in different groups. All data are expressed as the mean ± SD. aP < 0.05, bP < 0.01 compared with control group; cP < 0.05, dP < 0.01 compared with H2O2 group; eP < 0.05, fP < 0.01 compared with UA group. DHI: Disc height index; MRI: Magnetic resonance imaging; IDD: Intervertebral disc degeneration; UA: Urolithin A.
The degree of IVD degeneration was measured by MRI according to the Pfirrmann grade at 0 and 4 wk after puncture. As shown in Figure 7C-D, the Pfirrmann grade scores at 0 wk among the three groups did not show any significant difference (P > 0.05). However, the Pfirrmann grade scores of the IDD group were significantly higher than those of the control group, and the Pfirrmann grade scores of the UA group were lower than those of the IDD group at 4 wk (P < 0.01). The results indicated that UA intervention could alleviate IDD in vivo.
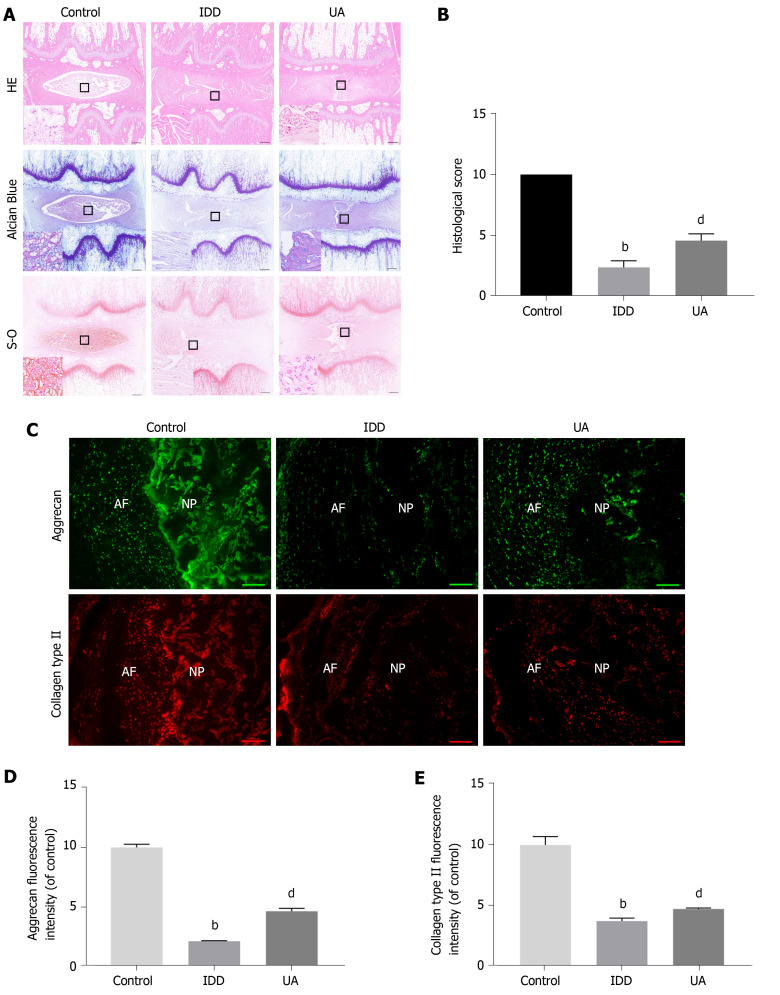
Histological analysis
As shown in Figure 8A-B, HE staining clearly showed inner well-structured gel-like NP and outer concentric ring-like annulus fibrosis, inner NP and cartilage endplates in the control group. In contrast, the well-structured IVD tissue was destroyed, and the NP tissue almost disappeared in the IDD group. However, a small number of NP cells and ECM still existed in the UA group. The histological score of the IDD group was also significantly lower than that of the control group, but UA treatment increased the histological score (P < 0.01).
Figure 8.
Hematoxylin-eosin, Safranin-O and Toluidine blue staining. A: Hematoxylin-eosin staining at 4 wk after puncture in different groups; B: Quantitative analysis of histological score in different groups (scale bar = 1 mm); C: The expression of aggrecan and collagen type II in different groups (scale bar = 200 μm); D-E: Quantitative analysis of aggrecan and collagen type II in different groups. All data are expressed as the mean ± SD. aP < 0.05, bP < 0.01 compared with control group; cP < 0.05, dP < 0.01 compared with H2O2 group; eP < 0.05, fP < 0.01 compared with UA group. HE: Hematoxylin-eosin; NP: Nucleus pulposus; AF: Annulus fibrosus; IDD: Intervertebral disc degeneration; UA: Urolithin A.
Safranin-O staining showed that the proteoglycan matrix (red positive tissue) level was significantly lower in the IDD group than in the control group. However, UA treatment protected the proteoglycan matrix from decreasing, and the proteoglycan matrix level in the UA group was higher than that of the IDD group. Similarly, toluidine blue staining revealed more NP chondrocytes in the UA group than in the IDD group.
Measurement of collagen type II and aggrecan
The expression of collagen type II and aggrecan in the disc tissue was assessed by immunofluorescence. As shown in Figure 8C-E, only a small portion of NP tissues showed positive expression of collagen type II and aggrecan in the IDD group, which were significantly lower than those of the control group (P < 0.01). However, the expression of collagen type II and aggrecan was higher than that of the IDD group after treatment with UA for 4 wk, which indicates that UA treatment can delay the downward trend of collagen type II and aggrecan (P < 0.05).
DISCUSSION
Our data demonstrated that UA could alleviate oxidative stress-induced NPMSCs senescence by activating the SIRT1/PGC-1α signaling pathway. NPMSCs harvested from the rat tails presented long spindle shapes and grew in flower formation. The ISCT for MCS has proposed the minimal criteria to define MSCs: (1) Plastic adherence characteristics; (2) Expression of CD105, CD73 and CD90 and lack expression of CD45, CD34, CD14 or CD11b, CD79a or CD19 and HLA-DR surface molecules; and (3) Multilineage differentiation potential to osteoblasts, adipocytes and chondroblasts in vitro[21]. Acquired NPMSCs were also found to be positive for CD105, CD73 and CD90 and negative for CD45 and CD34 through immunofluorescence and to successfully differentiate into osteogenic, chondrogenic and adipogenic differentiation. According to the above results, the cells isolated from the NP tissues met the criteria stated by the ISCT.
H2O2 is commonly used to induce oxidative damage in mechanistic studies of IDD[20,22]. The inflammation, apoptosis and senescence of NP cells and the apoptosis and senescence of NPMSCs are important pathological models for exploring IDD, which can all be induced by H2O2 treatment[23-26]. In our previous study, we confirmed that H2O2 treatment could decrease the viability of NPMSCs in a dose- and concentration-dependent manner, and a concentration of 80 μM for 6 h could be used as a suitable concentration in vitro[20]. Cell senescence often exhibits the characteristics of irreversible cell cycle arrest, loss of proliferation capacity and reduced cell anabolic ability. We evaluated the senescence of NPMSCs induced by H2O2 through SA-β-Gal staining, cell cycle, cell proliferation ability and cytotoxicity. The results demonstrated that an increased positive rate of SA-β-Gal staining and cytotoxicity, arrested the cell cycle and weakened the cell proliferation ability were found after H2O2 treatment. As special markers, the expression of p16 and p21 is particularly important to reflect cell senescence and the cell cycle[27,28]. After H2O2 treatment, the decreased expression of p16 and p21 further confirmed that oxidative stress can induce NPMSCs senescence.
As a metabolite of ellagitannin and ellagic acid, UA shows anti-inflammatory, antioxidant and antiaging effects[10,11,13]. To explore the effect of UA on the senescence of NPMSCs, the effect of different concentrations and different time points of UA treatment on cell activity was assessed through CCK-8, and the results showed that 20 μM can be used as an appropriate intervention concentration. Then, NPMSCs were pretreated with UA before H2O2 intervention, and the results showed that the cell proliferation capacity was restored, the percentage of SA-β-Gal staining positive cells and the cytotoxicity induced by H2O2 was less than that of the H2O2 group, which indicated that UA might protect NPMSCs against oxidative stress damage. Moreover, the number of cells arrested in the G2/M phase decreased by 11.72% after UA pretreatment, allowing more cells to enter a new cell cycle.
A previous study found that UA potently prolongs the lifespan of C. elegans by activating mitochondrial biogenesis and mitochondrial functions[13]. Normal mitochondrial function is essential for maintaining intracellular energy metabolism[29]. During the process of senescence, cells show an increased number of mitochondria and decreased membrane potential of these mitochondria. Decreased mitochondrial function results in the release of mitochondrial enzymes and overdose production of ROS[30]. By detecting the MMP of NPMSCs, we found that the content of J-aggregates of MMP after H2O2 treatment was significantly reduced (red fluorescence downregulated), while the content of J-aggregates was partly recovered after UA treatment. Thus, UA might have an antiaging effect by regulating the function of mitochondria. Similarly, Cásedas et al[31] investigated the antioxidative and neuroprotective effects of UA on the murine Neuro-2a neuroblastoma cell line and found that UA could improve mitochondrial activity, decrease lipid peroxidation and enhance the activity of antioxidant-related enzymes in cells subjected to oxidative stress. Excessive accumulation of intracellular ROS is also an important factor in cell senescence[32]. Oxidative stress leads to excessive ROS production, which further leads to DNA damage, protein damage and mitochondrial dysfunction[33]. The intracellular ROS content of NPMSCs was also decreased after UA pretreatment. Therefore, regulating mitochondrial function and reducing ROS production may be the main mechanisms by which UA exerts antiaging effects on NPMSCs.
As a classic pathway regulating mitochondrial function, the SIRT1/PGC-1α pathway has been proven to be involved in the regulation of multiple pathological processes, such as antiaging and oxidative stress[34,35]. SIRT1, a member of the NAD+-dependent Sir2 histone deacetylase family, has been reported to regulate mitochondrial function and reduce oxidative stress[36]. PGC-1α is a key regulator of mitochondrial biogenesis and function that can be activated by SIRT1 through deacetylation[37]. SIRT1/PGC-1α pathway activation attenuates oxidative damage and protects against metabolic disease, whereas the decreased activation of the SIRT1/PGC-1α axis is often closely related to some diseases characterized by mitochondrial disorders[35,37,38]. To address whether UA regulates mitochondrial function by activating the SIRT1/PGC-1α pathway to delay the senescence of NPMSCs, NPMSCs were treated together with the PGC-1α pathway inhibitors SR-18292 and UA. The results showed that the protective effect of UA was reversed by SR-18292. We also further evaluated the mRNA expression of SIRT1 and PGC-1α, and the results showed that the expression of SIRT1 and PGC-1α was significantly upregulated in the H2O2 + UA group compared with the H2O2 group. However, the mRNA expression levels of SIRT1 and PGC-1α were downregulated after SR-18292 treatment, which indicated that UA may have an antiaging effect by activating the SIRT1/PGC-1α pathway.
To evaluate the protective effect of UA more comprehensively, we also administered UA to IDD animal models. Delaying the loss of disc height and signal intensity of NP tissue also confirmed that UA can relieve IDD in vivo. Liu et al[15] found that UA treatment decreased matrix metalloproteinase production and the loss of collagen type II. We evaluated the expression of ECM at the histological level and found that UA can indeed delay the degradation of collagen type II and aggrecan, which further confirms that UA has a protective effect on degenerative IVD.
Admittedly, UA is reported to have pleiotropic properties, including the activation of signal pathways involving phosphatidylinositide 3-kinases, c-jun N-terminal kinase, nuclear factor-erythroid 2-related factor 2 and AMP-activated protein kinase[14,39,40]. However, in senescent NPMSCs induced by oxidative stress, we observed expression changes in pathway-related genes. Since we have not evaluated additional signaling pathways involved in the regulation of NPMSCs senescence, it is difficult to determine whether SIRT1/PGC-1α is the only pathway that regulates oxidative stress-induced NPMSCs senescence. Therefore, more signaling pathways that regulate the senescence of NPMSCs are worth exploring and discovering.
CONCLUSION
In summary, as shown in Figure 9, this study evaluated the protective effect of UA on oxidative stress-induced senescence in NPMSCs for the first time. H2O2 exposure could induce NPMSCs senescence and mitochondrial dysfunction. UA could activate the SIRT1/PGC-1α signaling pathway to protect mitochondrial function and alleviate cell senescence in vitro. UA could also delay ECM degradation and IDD in vivo. The results provide the possibility of promoting endogenous repair and retarding IDD.
Figure 9.
Schematic of protective effects of Urolithin A. Urolithin A activates the SIRT1/PGC-1α signaling pathway to protect mitochondrial function, alleviate nucleus pulposus-derived mesenchymal stem cells senescence in vitro, and delay intervertebral disc degeneration in vivo. SIRT1/PGC-1α: Silent information regulator of transcription 1/PPAR gamma coactivator-1α; NPMSCs: Nucleus pulposus-derived mesenchymal stem cells; UA: Urolithin A.
ARTICLE HIGHLIGHTS
Research background
Intervertebral disc degeneration (IDD) is the main pathogenic factor of low back pain, but its pathological mechanism has not yet been elucidated. The isolation and identification of nucleus pulposus-derived mesenchymal stem cells (NPMSCs) provided a basis for the endogenous repair of IDD.
Research motivation
An unfavorable microenvironment of degenerative intervertebral disc such as inflammation, oxidative stress, and increased catabolism leads to increased senescence NPMSCs, which seriously affects endogenous repair. Therefore, rescuing the activity of NPMSCs and delaying cell senescence is of great significance to alleviate IDD.
Research objectives
The present study investigated whether urolithin A (UA) could alleviate NPMSCs senescence induced by oxidative stress and the potential mechanism.
Research methods
The protective effects of UA against oxidative stress-induced senescence in NPMSCs were investigated by evaluating the senescence-associated β-Galactosidase (SA-β-Gal) activity, cell cycle, cell proliferation ability, mitochondrial function and reactive oxygen species (ROS). Additionally, the expression of senescence-related and the silent information regulator of transcription 1/PPAR gamma coactivator-1α (SIRT1/PGC-1α) pathway-related proteins and mRNA was also used to evaluate the protective effects of UA in vitro. In vivo, an animal model of IDD were constructed, and X-rays, magnetic resonance imaging, and histological analysis were used to assessed whether UA could alleviate IDD in vivo.
Research results
in vitro, UA could reduce SA-β-Gal activity and senescence-related proteins and mRNA (P16 and P21) expression, alleviate cell cycle arrest and ROS production, stimulate cell proliferation ability and mitochondrial function by activating the SIRT1/PGC-1α pathway. In vivo, UA could alleviate an animal model of IDD by assessed the disc height index, Pfirrmann grade and the histological score.
Research conclusions
UA could activate the SIRT1/PGC-1α signaling pathway to protect mitochondrial function and alleviate cell senescence, and further delay extracellular matrix degradation and IDD, which provide the possibility of promoting endogenous repair and retarding IDD.
Research perspectives
We demonstrated the positive role of UA in attenuating oxidative stress-induced NPMSCs senescence and delaying IDD. UA may be successfully applied to IDD endogenous repair.
Footnotes
Institutional review board statement: This study was approved by the Ethical Committee of the Clinical Medical College of Yangzhou University (SBYY2020-023).
Institutional animal care and use committee statement: All animal experiments conformed to the internationally accepted principles for the care and use of laboratory animals (Shanghai Institute of Family Planning Science, License No. SCXK (Hu) 2018-0006).
Conflict-of-interest statement: The authors have no relevant financial or non-financial interests to disclose.
Data sharing statement: No additional data are available.
ARRIVE guidelines statement: The authors have read the ARRIVE guidelines, and the manuscript was prepared and revised according to the ARRIVE guidelines.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: National Natural Science Foundation of China, 81972136; Young Medical Scholars Major Program of Jiangsu Province, QNRC2016342; Key Funding Project of Maternal and Child Health Research of Jiangsu Province, F201801; High-level Health Professionals "Six projects" Top-notch Talent Research Program of Jiangsu Province, LGY2019035.
Peer-review started: July 2, 2021
First decision: July 29, 2021
Article in press: November 28, 2021
Specialty type: Orthopedics
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mogulkoc R, Zhang M S-Editor: Liu M L-Editor: A P-Editor: Liu M
Contributor Information
Peng-Zhi Shi, Department of Orthopedic, Dalian Medical University, Dalian 116000, Liaoning Province, China.
Jun-Wu Wang, Department of Orthopedics, Clinical Medical College of Yangzhou University, Yangzhou 225000, Jiangsu Province, China.
Ping-Chuan Wang, Department of Orthopedics, Clinical Medical College of Yangzhou University, Yangzhou 225000, Jiangsu Province, China.
Bo Han, Department of Orthopedic, Beijing Chaoyang Hospital, Capital Medical University, Beijing 100020, China.
Xu-Hua Lu, Department of Orthopedics, Changzheng Hospital of The Second Military Medical University, Shanghai 200003, China.
Yong-Xin Ren, Department of Orthopedics, First Affiliated Hospital of Nanjing Medical University, Nanjing 210029, Jiangsu Province, China.
Xin-Min Feng, Department of Orthopedics, Clinical Medical College of Yangzhou University, Yangzhou 225000, Jiangsu Province, China.
Xiao-Fei Cheng, Department of Orthopedic Surgery, Shanghai Key Laboratory of Orthopedics Implants, Shanghai Ninth People’s Hospital, Shanghai JiaoTong University School of Medicine, Shanghai 200011, China.
Liang Zhang, Department of Orthopedics, Clinical Medical College of Yangzhou University, Yangzhou 225000, Jiangsu Province, China. zhangliang6320@sina.com.
References
- 1.Dagenais S, Caro J, Haldeman S. A systematic review of low back pain cost of illness studies in the United States and internationally. Spine J . 2008;8:8–20. doi: 10.1016/j.spinee.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Manchikanti L, Singh V, Falco FJ, Benyamin RM, Hirsch JA. Epidemiology of low back pain in adults. Neuromodulation . 2014;17 Suppl 2:3–10. doi: 10.1111/ner.12018. [DOI] [PubMed] [Google Scholar]
- 3.Zheng CJ, Chen J. Disc degeneration implies low back pain. Theor Biol Med Model . 2015;12:24. doi: 10.1186/s12976-015-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, Peroglio M, Alini M, Grad S. Potential and limitations of intervertebral disc endogenous repair. Curr Stem Cell Res Ther . 2015;10:329–338. doi: 10.2174/1574888x10666150305105114. [DOI] [PubMed] [Google Scholar]
- 5.Grad S, Peroglio M, Li Z, Alini M. Endogenous cell homing for intervertebral disk regeneration. J Am Acad Orthop Surg . 2015;23:264–266. doi: 10.5435/JAAOS-D-15-00096. [DOI] [PubMed] [Google Scholar]
- 6.Risbud MV, Guttapalli A, Tsai TT, Lee JY, Danielson KG, Vaccaro AR, Albert TJ, Gazit Z, Gazit D, Shapiro IM. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine (Phila Pa 1976) . 2007;32:2537–2544. doi: 10.1097/BRS.0b013e318158dea6. [DOI] [PubMed] [Google Scholar]
- 7.Vadalà G, Ambrosio L, Russo F, Papalia R, Denaro V. Interaction between Mesenchymal Stem Cells and Intervertebral Disc Microenvironment: From Cell Therapy to Tissue Engineering. Stem Cells Int . 2019;2019:2376172. doi: 10.1155/2019/2376172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomás-Barberán FA, González-Sarrías A, García-Villalba R, Núñez-Sánchez MA, Selma MV, García-Conesa MT, Espín JC. Urolithins, the rescue of "old" metabolites to understand a "new" concept: Metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol Nutr Food Res . 2017;61 doi: 10.1002/mnfr.201500901. [DOI] [PubMed] [Google Scholar]
- 9.Cerdá B, Llorach R, Cerón JJ, Espín JC, Tomás-Barberán FA. Evaluation of the bioavailability and metabolism in the rat of punicalagin, an antioxidant polyphenol from pomegranate juice. Eur J Nutr . 2003;42:18–28. doi: 10.1007/s00394-003-0396-4. [DOI] [PubMed] [Google Scholar]
- 10.Bialonska D, Kasimsetty SG, Khan SI, Ferreira D. Urolithins, intestinal microbial metabolites of Pomegranate ellagitannins, exhibit potent antioxidant activity in a cell-based assay. J Agric Food Chem . 2009;57:10181–10186. doi: 10.1021/jf9025794. [DOI] [PubMed] [Google Scholar]
- 11.Boakye YD, Groyer L, Heiss EH. An increased autophagic flux contributes to the anti-inflammatory potential of urolithin A in macrophages. Biochim Biophys Acta Gen Subj . 2018;1862:61–70. doi: 10.1016/j.bbagen.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Chen JH, Aguilera-Barrantes I, Shiau CW, Sheng X, Wang LS, Stoner GD, Huang YW. Urolithin A suppresses the proliferation of endometrial cancer cells by mediating estrogen receptor-α-dependent gene expression. Mol Nutr Food Res . 2016;60:2387–2395. doi: 10.1002/mnfr.201600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Félix AA, Williams EG, Jha P, Lo Sasso G, Huzard D, Aebischer P, Sandi C, Rinsch C, Auwerx J. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med . 2016;22:879–888. doi: 10.1038/nm.4132. [DOI] [PubMed] [Google Scholar]
- 14.Lin J, Zhuge J, Zheng X, Wu Y, Zhang Z, Xu T, Meftah Z, Xu H, Tian N, Gao W, Zhou Y, Zhang X, Wang X. Urolithin A-induced mitophagy suppresses apoptosis and attenuates intervertebral disc degeneration via the AMPK signaling pathway. Free Radic Biol Med . 2020;150:109–119. doi: 10.1016/j.freeradbiomed.2020.02.024. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Kang H, Song C, Lei Z, Li L, Guo J, Xu Y, Guan H, Fang Z, Li F. Urolithin A Inhibits the Catabolic Effect of TNFα on Nucleus Pulposus Cell and Alleviates Intervertebral Disc Degeneration in vivo. Front Pharmacol . 2018;9:1043. doi: 10.3389/fphar.2018.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T, Cheng X, Wang J, Feng X, Zhang L. Time-Course Investigation of Intervertebral Disc Degeneration Induced by Different Sizes of Needle Punctures in Rat Tail Disc. Med Sci Monit . 2018;24:6456–6465. doi: 10.12659/MSM.910636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han B, Zhu K, Li FC, Xiao YX, Feng J, Shi ZL, Lin M, Wang J, Chen QX. A simple disc degeneration model induced by percutaneous needle puncture in the rat tail. Spine (Phila Pa 1976) . 2008;33:1925–1934. doi: 10.1097/BRS.0b013e31817c64a9. [DOI] [PubMed] [Google Scholar]
- 18.Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) . 2001;26:1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 19.Norcross JP, Lester GE, Weinhold P, Dahners LE. An in vivo model of degenerative disc disease. J Orthop Res . 2003;21:183–188. doi: 10.1016/S0736-0266(02)00098-0. [DOI] [PubMed] [Google Scholar]
- 20.Nan LP, Wang F, Liu Y, Wu Z, Feng XM, Liu JJ, Zhang L. 6-gingerol protects nucleus pulposus-derived mesenchymal stem cells from oxidative injury by activating autophagy. World J Stem Cells . 2020;12:1603–1622. doi: 10.4252/wjsc.v12.i12.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz M, Cosenza S, Maumus M, Jorgensen C, Noël D. Therapeutic application of mesenchymal stem cells in osteoarthritis. Expert Opin Biol Ther . 2016;16:33–42. doi: 10.1517/14712598.2016.1093108. [DOI] [PubMed] [Google Scholar]
- 22.Dimozi A, Mavrogonatou E, Sklirou A, Kletsas D. Oxidative stress inhibits the proliferation, induces premature senescence and promotes a catabolic phenotype in human nucleus pulposus intervertebral disc cells. Eur Cell Mater . 2015;30:89–102; discussion 103. doi: 10.22203/ecm.v030a07. [DOI] [PubMed] [Google Scholar]
- 23.Nan LP, Wang F, Ran D, Zhou SF, Liu Y, Zhang Z, Huang ZN, Wang ZY, Wang JC, Feng XM, Zhang L. Naringin alleviates H2O2-induced apoptosis via the PI3K/Akt pathway in rat nucleus pulposus-derived mesenchymal stem cells. Connect Tissue Res . 2020;61:554–567. doi: 10.1080/03008207.2019.1631299. [DOI] [PubMed] [Google Scholar]
- 24.Xia C, Zeng Z, Fang B, Tao M, Gu C, Zheng L, Wang Y, Shi Y, Fang C, Mei S, Chen Q, Zhao J, Lin X, Fan S, Jin Y, Chen P. Mesenchymal stem cell-derived exosomes ameliorate intervertebral disc degeneration via anti-oxidant and anti-inflammatory effects. Free Radic Biol Med . 2019;143:1–15. doi: 10.1016/j.freeradbiomed.2019.07.026. [DOI] [PubMed] [Google Scholar]
- 25.Tang P, Gu JM, Xie ZA, Gu Y, Jie ZW, Huang KM, Wang JY, Fan SW, Jiang XS, Hu ZJ. Honokiol alleviates the degeneration of intervertebral disc via suppressing the activation of TXNIP-NLRP3 inflammasome signal pathway. Free Radic Biol Med . 2018;120:368–379. doi: 10.1016/j.freeradbiomed.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Lin J, Du J, Wu X, Xu C, Liu J, Jiang L, Cheng X, Ge G, Chen L, Pang Q, Geng D, Mao H. SIRT3 mitigates intervertebral disc degeneration by delaying oxidative stress-induced senescence of nucleus pulposus cells. J Cell Physiol . 2021;236:6441–6456. doi: 10.1002/jcp.30319. [DOI] [PubMed] [Google Scholar]
- 27.Demaria M, Ohtani N, Youssef SA, Rodier F, Toussaint W, Mitchell JR, Laberge RM, Vijg J, Van Steeg H, Dollé ME, Hoeijmakers JH, de Bruin A, Hara E, Campisi J. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell . 2014;31:722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal . 2010;22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature . 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of Cellular Senescence. Trends Cell Biol . 2018;28:436–453. doi: 10.1016/j.tcb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Cásedas G, Les F, Choya-Foces C, Hugo M, López V. The Metabolite Urolithin-A Ameliorates Oxidative Stress in Neuro-2a Cells, Becoming a Potential Neuroprotective Agent. Antioxidants (Basel) . 2020;9:177. doi: 10.3390/antiox9020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol . 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Wu Q, Wang Y, Li L, Bu H, Bao J. Senescence of mesenchymal stem cells (Review) Int J Mol Med . 2017;39:775–782. doi: 10.3892/ijmm.2017.2912. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Feng L, Xing Y, Wang Y, Du L, Xu C, Cao J, Wang Q, Fan S, Liu Q, Fan F. Radioprotective and antioxidant effect of resveratrol in hippocampus by activating Sirt1. Int J Mol Sci . 2014;15:5928–5939. doi: 10.3390/ijms15045928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang D, Zhuo Y, Guo Z, He L, Wang X, He Y, Li L, Dai H. SIRT1/PGC-1 pathway activation triggers autophagy/mitophagy and attenuates oxidative damage in intestinal epithelial cells. Biochimie . 2020;170:10–20. doi: 10.1016/j.biochi.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells . 2014;32:1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell . 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell . 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komatsu W, Kishi H, Yagasaki K, Ohhira S. Urolithin A attenuates pro-inflammatory mediator production by suppressing PI3-K/Akt/NF-κB and JNK/AP-1 signaling pathways in lipopolysaccharide-stimulated RAW264 macrophages: Possible involvement of NADPH oxidase-derived reactive oxygen species. Eur J Pharmacol . 2018;833:411–424. doi: 10.1016/j.ejphar.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 40.Singh R, Chandrashekharappa S, Bodduluri SR, Baby BV, Hegde B, Kotla NG, Hiwale AA, Saiyed T, Patel P, Vijay-Kumar M, Langille MGI, Douglas GM, Cheng X, Rouchka EC, Waigel SJ, Dryden GW, Alatassi H, Zhang HG, Haribabu B, Vemula PK, Jala VR. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun . 2019;10:89. doi: 10.1038/s41467-018-07859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]