Fig. 3.
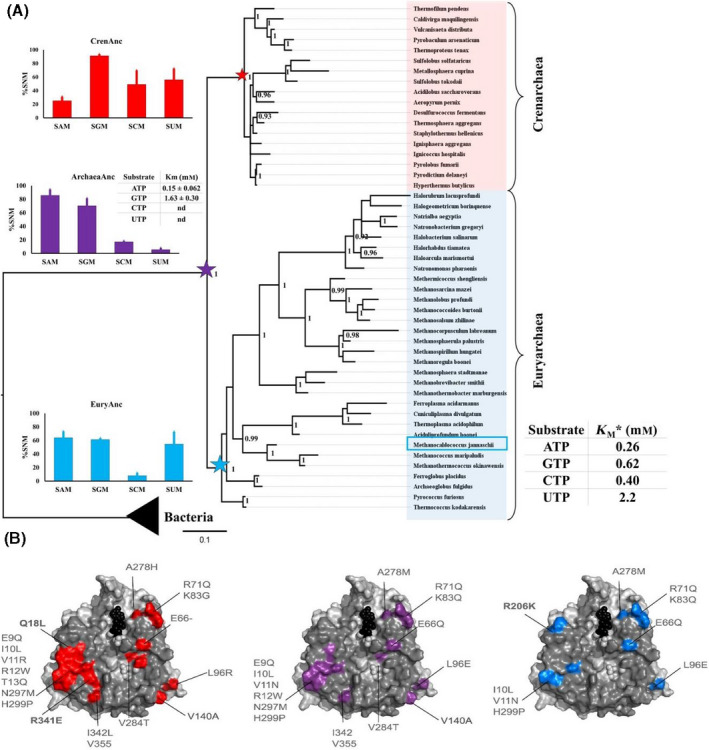
Phylogenetic tree of archaeal MAT with bacteria domain as outgroup and activity assays of the inferred ancestors. (A) The maximum‐likelihood phylogenetic tree topology of archaeal MAT with bifurcation into two major clades, that is, Crenarchaea and EuryAnc, and with bacteria as an outgroup. This topology is further supported by another phylogenetic tree based on the 16s rRNA sequences from the corresponding species. Here, *we highlight the activity of MjMAT as representative of domain archaea, with the data as reported previously [48]. Ancestral sequences were inferred for the ancestor of Crenarchaea MAT (CrenArc in red star), EuryAnc MAT (EuryArc in blue star), and the common ancestor for the archaeal MAT (ArcheaArc in violet star). Bayesian posterior probability values > 90% are shown. One time point assay for ancestral sequence activities was performed with corresponding enzyme (20 µm) using NTPs (5 mm) and methionine (10 mm) in HEPES (100 mm), KCl (50 mm), and MgCl2(10 mm), pH 8, at 37 °C for hMAT1A and 55 °C for archaea, 1 h. For kinetics analysis, ArcheaArc MAT (0.5 µm), ATP, and GTP (0.1–2 mm) and methionine (10 mm) using the same above‐mentioned buffer and conditions were used. The experiments were performed in duplicates. The error bars represent SD. Tables show the extrapolated K M for ArcheaArc. The production of SAM and SGM was analyzed by UPLC, and data were fitted to the Michaelis–Menten equation using graphpad software 7.02. (B) Mutations found by the ancestral reconstruction analysis are highlighted on the available structure of pfMAT (PDB: 6S83). The structures are represented as gray surface and show the large interface. CrenAnc MAT interface mutations are highlighted in red, EuryAnc in blue, and ArchaeaAnc in violet.
