Fig. 5.
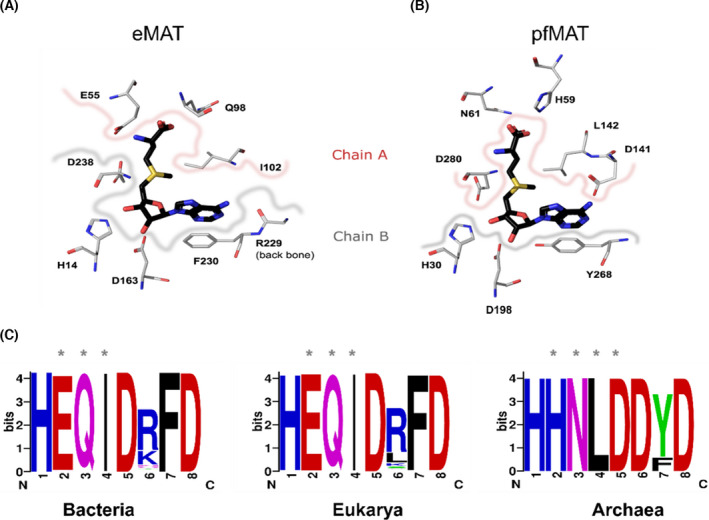
Comparison of the active site residues in the different domains. (A) Active site residues involved in the interaction with SAM. SAM is highlighted in black, while the active site residues from eMAT (PDB: 1RG9) have been highlighted in metal gray. The residues derived from chain A (E55, Q98, I102) have been demarcated with a pinkish border, and residues from chain B (H14, D163, R229, F230, D238) have been demarcated with a grayish border. (B) Equivalent active site residues from pfMAT [33] (PDB: 6S83) that interact with SAM have also been highlighted from chain A (H59, N61, D141, L142, D280) and chain B (H30, D198, Y268) using the same color codes. (C) The active site residues identified based on structural alignment, from all three domains of life, have been highlighted as WebLogos (https://weblogo.berkeley.edu/logo.cgi). Residues derived from the second monomeric subunit have been marked using ‘*’. WebLogo was generated using alignment of 526 sequences from bacteria (Appendix S1), 273 sequences from eukarya (Appendix S2), and 49 sequences from archaea (Appendix S3). Details of sequence search are in the Methods section.
