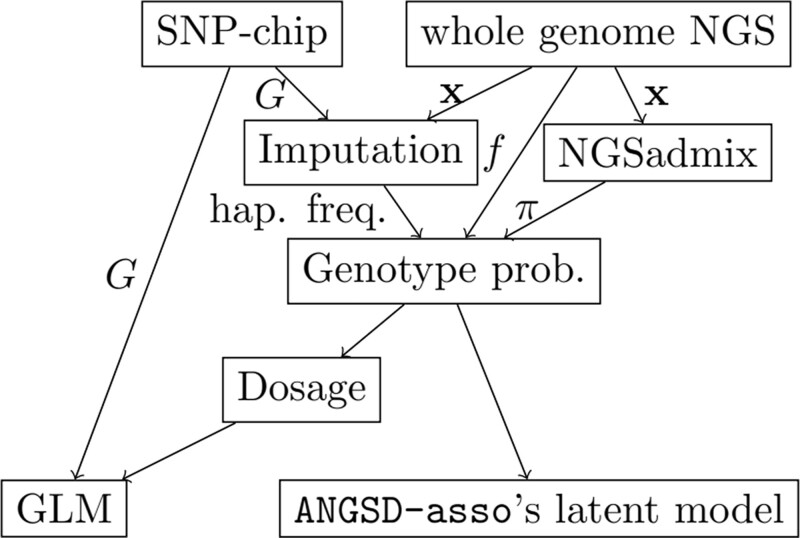
Figure 1.
Schematic of workflow for performing association studies with genetic data. x is the sequence data that can be converted to genotype likelihoods, G is the genotypes. π is the individual allele frequencies and f is the sample allele frequency both of which can be used as priors when estimating the genotype probabilities. Data either gets generated using SNP-chips or doing NGS. The NGS data can be converted into genotype probabilities assuming no population structure using the sample allele frequency, or assuming population structure and then using principal component analysis (PCA) to generate genotype probabilities. SNP-chip genotypes can be analyzed directly. Both kinds of data can be imputed using haplotype frequencies for generating genotype probabilities. The genotype probabilities can be analyzed with ANGSD-asso’s latent model or converted to dosages and then be analyzed with a GLM.