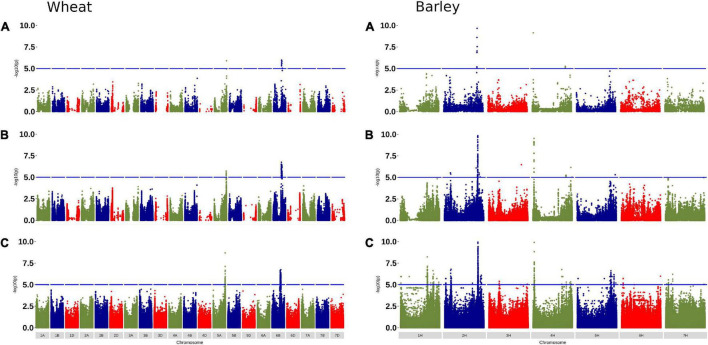
FIGURE 5.
Genome-wide association study (GWAS) for awned status and row type in wheat and barley, respectively, using: (A) selected tSNP; (B) selected tSNP after imputation to the r2 ≥ 0.7 target set (the sample being imputed was removed from the reference set); and (C) exome SNP. Note the -log10(p) axes are scaled to 10 which resulted in the most significant SNP (38.44) for the 5A locus in wheat being out of the range on the axis for the wheat exome SNP plot.