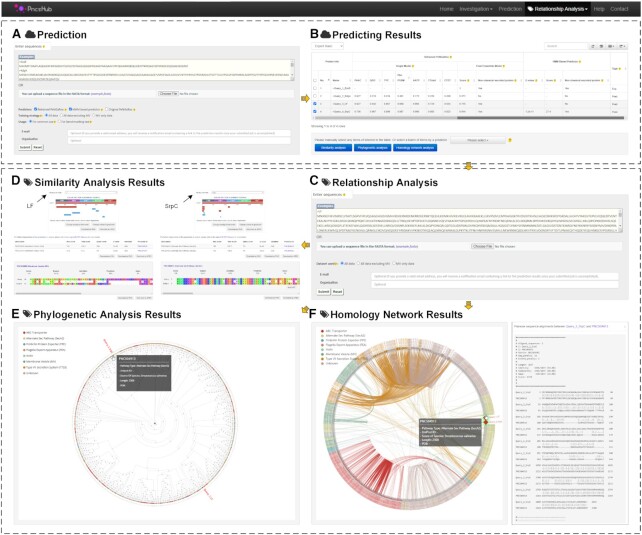
Figure 4.
Graphical illustration of the functional modules in PncsHub. (A, B) Example sequences were submitted as query proteins to both the HMM and retrained PeNGaRoo prediction tools: EsxB (PNCS00367), LF (PNCS00598), SrpC (PNCS04914), and the negative control KdgA (UniProt ID: P50846). EsxB, LF, and SrpC are experimentally validated non-classically secreted substrates (not included into any of our training datasets), but only EsxB is included in the ‘filter list’. (C) LF and SrpC were subsequently transferred to our Relationship Analysis Module to determine likely secretion pathways using (D) Similarity Analysis, (E) Phylogenetic Analysis, and (F) Homology Network Analysis.