Figure 1.
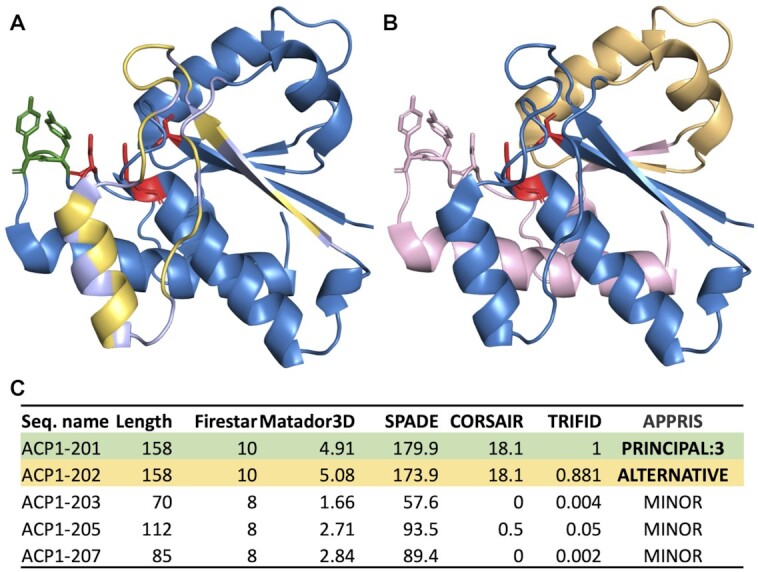
APPRIS annotations for ACP1 from the GENCODE v38 reference set. Panels A and B show PDB structure 4Z99, the resolved structure of isoform A of low molecular weight protein tyrosine phosphatase (ACP1-001 in the GENCODE annotation), onto which has been mapped the effects of alternative splicing for two different annotated isoforms, ACP1-002 and ACP1-005. Known catalytic residues are shown as red sticks, Src-phosphorylated tyrosines are shown in green. Panel A shows variant ACP1-002, which differs from ACP1-001 by a single tandem duplicated homologous exon. Residues identical between ACP1-001 and ACP1-002 in the exon are shown in light blue; those that differ are shown in light yellow. Catalytic and phosphorylated residues are not affected by this exon. Panel B shows variant ACP1-005. In this case the second tandem duplicated exon is read in a different frame leading to a premature stop codon. The unrelated sequence from the frameshifted amino acids is mapped onto the structure in light orange, but these residues are likely to be unfolded. The remaining protein structure of ACP1-001 (shown in light pink) would be lost in this isoform, eliminating one of the catalytic residues and both phosphorylated tyrosines. Isoforms generated from ACP1-003 and ACP1-007 swap even more of the C-terminal region of the principal isoform for unrelated residues and premature stop codons. Both lose the same catalytic and phosphorylated residues as ACP1-005. Panel C shows the scores from the APPRIS modules for the five isoforms. The principal isoform scores are shown with a green background, the ‘alternative’ isoform with an orange background. The scores for variant ACP1-002 are so similar to those of the principal isoform that APPRIS has to determine the principal isoform from external methods. ACP1-001 is chosen as the principal isoform on the strength of proteomics evidence (PRINCIPAL:3). TRIFID predicts that ‘ALTERNATIVE’ variant ACP1-002 is highly likely to be functionally important as a protein. The remaining three isoforms lose protein structure, functional domains and residues and have no detectable cross-species conservation. TRIFID predicts that they will not be functionally relevant at the protein level.
