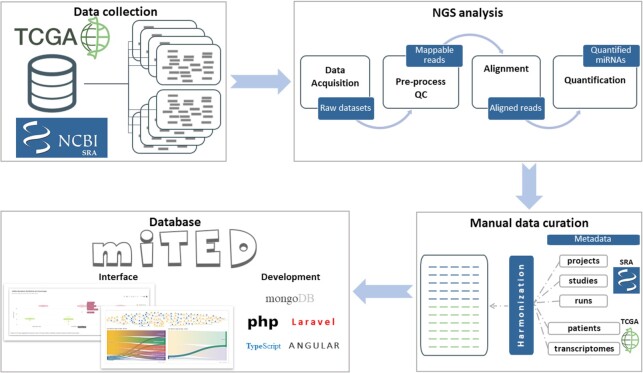
Figure 1.
DIANA-miTED development workflow. Initially, human raw sRNA-Seq datasets were retrieved from NCBI-SRA and TCGA (alignment files obtained from TCGA and converted back to FASTQ format). Raw datasets were uniformly subjected to pre-processing and quality control, alignment and quantification. Read count, RPM and log2(RPM) values were calculated for miRBase miRNAs. Metadata from both resources were curated manually to create a comprehensive, standardized set of metadata annotations for the analyzed datasets. DIANA-miTED resource was developed utilizing MongoDB (noSQL database), PHP/Laravel for data access layer development, and Typescript/Angular for the application layer development. miTED features extensive query, filtering and visualization options and supports local retrieval of requested data.