Figure 6.
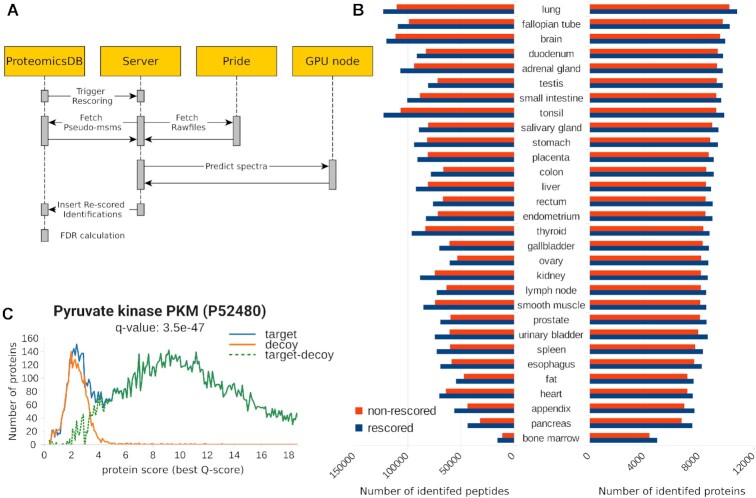
Integration of Prosit into ProteomicsDB. (A) Depiction of the workflow implemented to enable automatic rescoring of projects in ProteomicsDB. Raw mass spectrometry data are downloaded from PRIDE. The rescoring is performed on the database search results stored in ProteomicsDB by retrieving predictions from Prosit. The resulting scores are merged by percolator and imported into ProteomicsDB where the picked protein approach is used for FDR estimation. (B) The number of proteins (right) and peptides (left) identified with (blue) and without (red) rescoring at an estimated PSM, peptide and protein FDR of 1% for 30 tissues from Wang et al. (29). (C) Distribution of target and decoy Q-scores of proteins supported by peptide identifications for all mouse proteins in ProteomicsDB. The example highlights the q-value of the Pyruvate kinase PKM (P52480).