Figure 2.
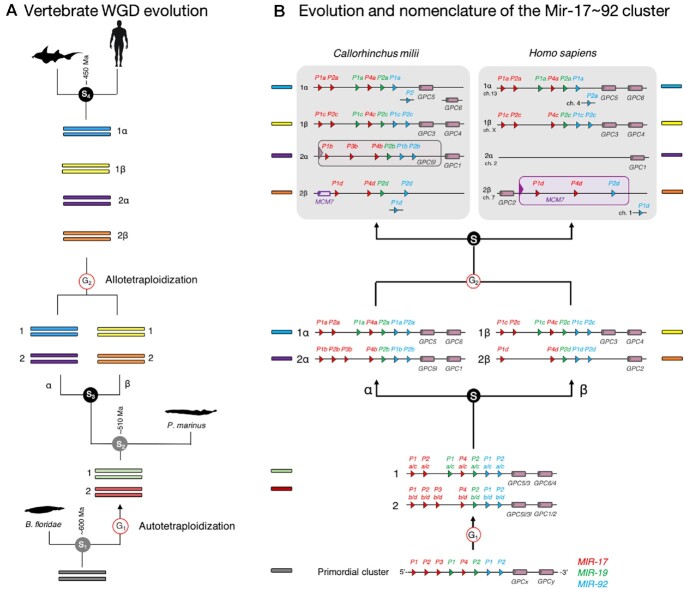
MirGeneDB nomenclature system of gnathostome microRNA genes. (A) Model of vertebrate genome evolution adapted from (43). The diploid state of an early chordate ancestor doubled in content (G1) through an autotetraploidy event (38) generating a tetraploid genome. Then, two lineages were generated from a speciation event (S3), α and β. Two species of these lineages then hybridized in an allotetraploidy event (G2), resulting in a single species with an octoploid genome. Sometime soon after this event, around 450 million years ago, the gnathostome LCA evolved and gave rise to the two major extant gnathostome lineages, the Chondrichthyes (the cartilaginous fish) and the Osteichthyes (the bony fish) (S4). (B) microRNA gene nomenclature of paralogues, orthologues, ohnologues (genes generated by autotetraploidy events, in this case sub-genomes 1 and 2) and homeologues (gene generated by allotetraploidy events, in this case paralogons α and β) as exemplified by the Mir-17∼92 cluster. See text for details.