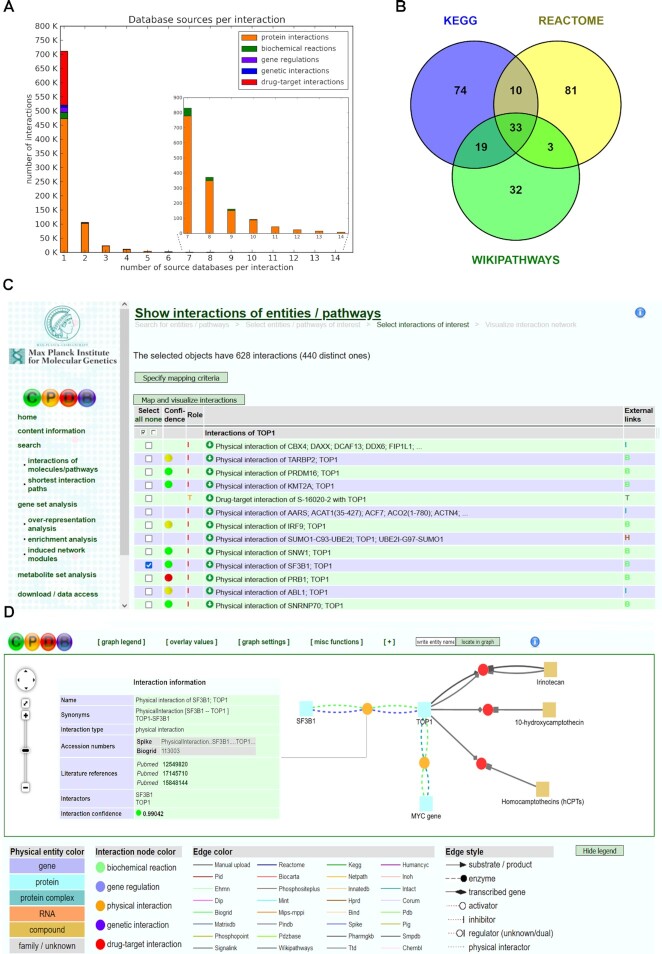
Figure 1.
ConsensusPathDB content. (A) Number of interactions (Y-axis) shared by number of source databases (X-axis). The rightmost tail of the histogram is magnified. The colors within each bar represent the different interaction types. (B) Venn diagram of overlapping gene sets for the apoptosis pathway annotated by three prominent pathway databases: KEGG (pathway identifier: hsa04210), Reactome (R-HSA-109581) and WikiPathways (WP254). (C) Interaction display for the TOP1 (DNA topoisomerase I) gene. Binary interactions are scored for confidence and the confidence value is displayed by a traffic light symbol. Each interaction assigns a specific role to the molecule under study (e.g. ‘I’ interactor, ‘T’ target) and has an external link to the annotating source database. Interactions can be selected for further visualization. (D) Visualization of selected interactions of TOP1. Interactions are displayed with colored nodes indicating the interaction type and interacting molecules are displayed with colored squares indicating their type. Each connection represents a source database that has annotated the interaction. Clicking on each interaction (or molecule) displays further information about the interaction, the confidence score and supporting publications.