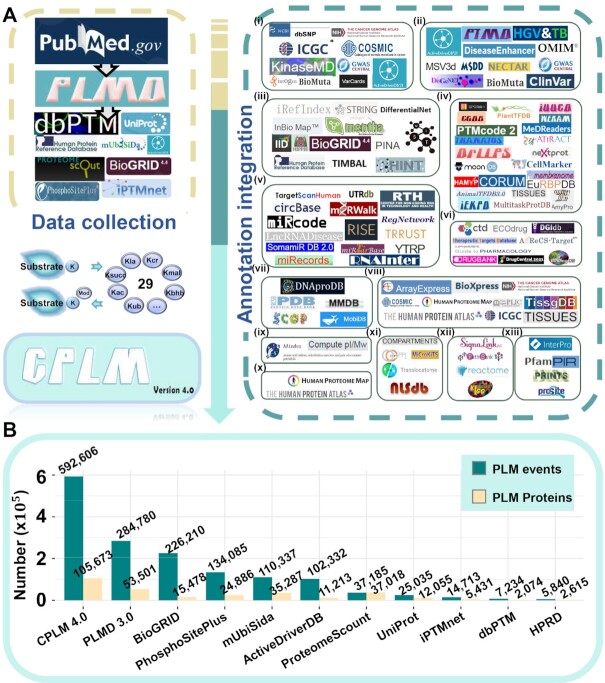
Figure 1.
The procedure for development of CPLM 4.0. (A) First, we manually collected experimentally identified PLM substrates and sites from PubMed. We also integrated the existing data of 10 public databases, including PLMD 3.0 (28), dbPTM (30), ProteomeScout (31), iPTMnet (32), BioGRID (33), PhosphoSitePlus (34), mUbiSiDa (35), HPRD (36), ActiveDriverDB (37) and UniProt (29) (Supplementary Table S1). Furthermore, we annotated the PLM proteins and sites, using the knowledge from 102 additional databases that covered 13 aspects: (i) variation and mutation; (ii) disease-associated information; (iii) protein–protein interaction; (iv) protein function; (v) DNA & RNA element; (vi) chemical–target relation; (vii) protein structure; (viii) mRNA expression; (ix) physicochemical property; (x) protein expression/proteomics; (xi) subcellular localization; (xii) biological pathway; (xiii) domain annotation (Supplementary Table S2). Kla, lysine lactylation; Kcr, lysine crotonylation; Kmal, lysine malonylation; Kbhb, lysine β-hydroxybutyrylation; Kub: lysine ubiquitination; Kac, lysine acetylation; Ksucc, lysine succinylation. (B) A comparison of PLM events and proteins between CPLM 4.0 and other existing resources.