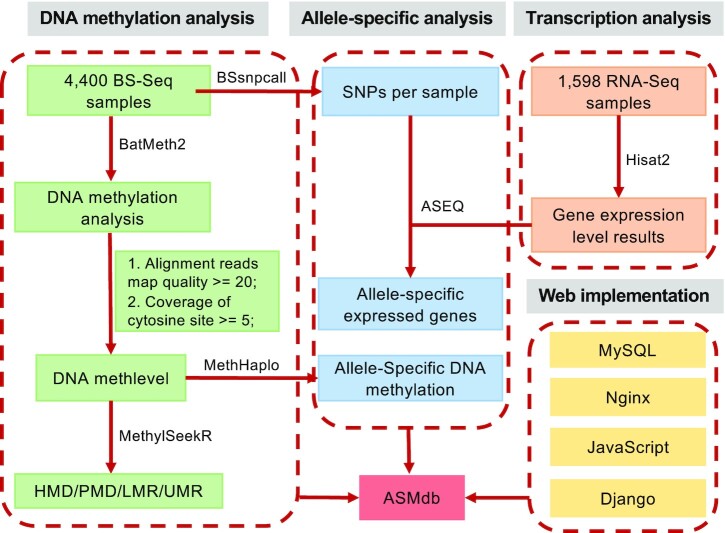
Figure 1.
Procedure used for ASMdb construction. The ASMdb database was constructed with MySQL and Django tools. BatMeth2 was used to map BS-Seq data, calculate the DNA methylation level and visualize the methylation patterns. MethHaplo was used to detect allele-specific DNA methylation. Hisat2 was used for RNA-Seq data mapping. ASEQ was used to detect allele-specific expressed genes. For annotation purposes, we used MethylSeekR to divide the genome into four categories of regions: unmethylated regions (UMRs), low-methylation regions (LMRs), partially methylated domains (PMDs) and highly methylated domains (HMDs) according to the methylation level.