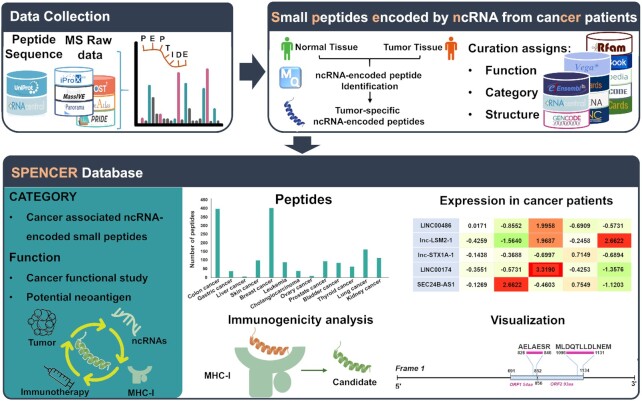
Figure 1.
Overall design and construction of SPENCER. From published literature and available databases, we collected ∼450 000 ncRNA sequences and ∼60 MS datasets in SPENCER (upper left). MaxQuant was applied to search for ncPEPs in the collected MS data. Meanwhile, differential expression analysis of the identified small peptides between tumors and normal/paracancerous tissues was performed. To investigate the potential roles of small peptides in cancer, SPENCER integrates detailed annotation of the associated ncRNAs with information from external resources, including function, category and structure (upper right). In addition, using a robust pipeline, the immunogenicity of all the identified ncPEPs was predicted based on three feature scores. Finally, we integrated and visualized the data obtained above to build the SPENCER database (lower).