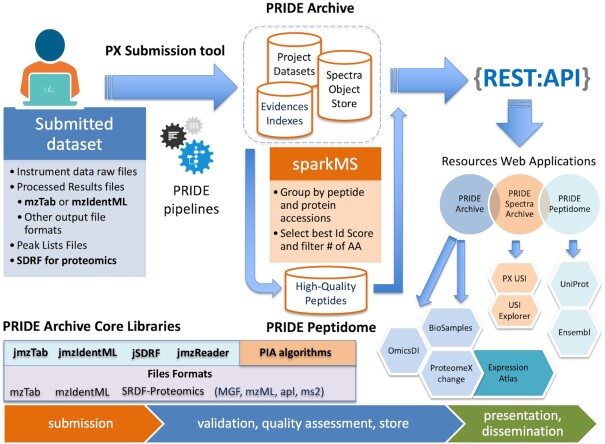
Figure 1.
Schema of the PRIDE resources ecosystem. PRIDE Archive users must provide the raw files, the processed results files, and metadata about every given dataset. Standard file formats (for processed result files) can be provided for 'Complete' submissions. A group of open-source libraries is used by the PX Submission tool, and the PRIDE pipelines to validate, assess the quality of the reported peptides and proteins, and store the information (metadata, peptides/proteins and spectra) into multiple databases. The PRIDE Peptidome resource selects high-quality peptides across all the datasets in PRIDE Archive. All the data from PRIDE Archive and PRIDE Peptidome is served to external users such as Ensembl and UniProt through the PRIDE API and PRIDE web interface. Additionally, proteomics quantitative datasets are reanalyzed and integrated into Expression Atlas.