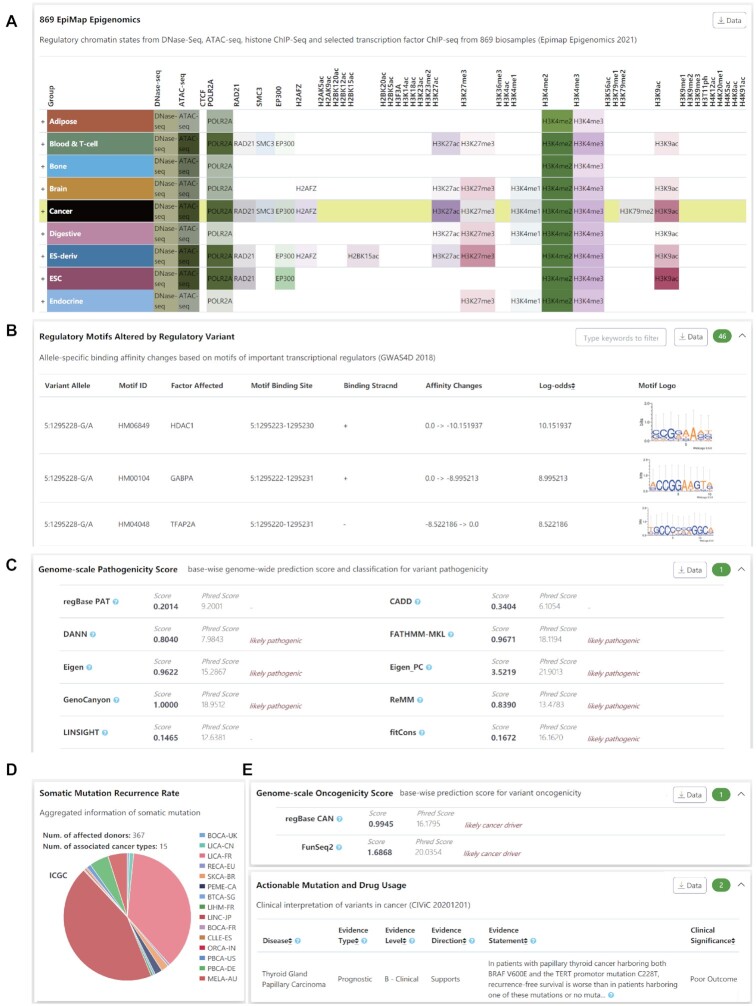
Figure 3.
Supporting evidence from VannoPortal for the regulatory potential and cancer-driven roles of chr5:g.1295228:G > A (GRCh37, rs1242535815). (A) rs1242535815 overlaps active chromatin states (e.g. DNase-seq and ATAC-seq), histone marks (e.g. H3K27ac, H3K4me2, H3K4me3 and H3K9ac) and TF binding sites (e.g. POLR2A, RAD21 and SMC3) across many human tissues, particularly in cancers. (B) rs1242535815 A allele potentially increases the binding affinity of HDAC1 and GABPA. (C) rs1242535815 is a likely pathogenic mutation supported by many genome-scale base-wise pathogenicity prediction methods. (D) rs1242535815 is a highly recurrent mutation in cancer patients. (E) rs1242535815 is a likely cancer driver mutation supported by regBase-CAN and other tools, and it could be a prognostic marker in cancer therapy.