Figure 1. Validation of SNRPN Promoter Methylation Analysis Using Methylation Specific Quantitative Melt Analysis (MS-QMA) on 1356 Samples.
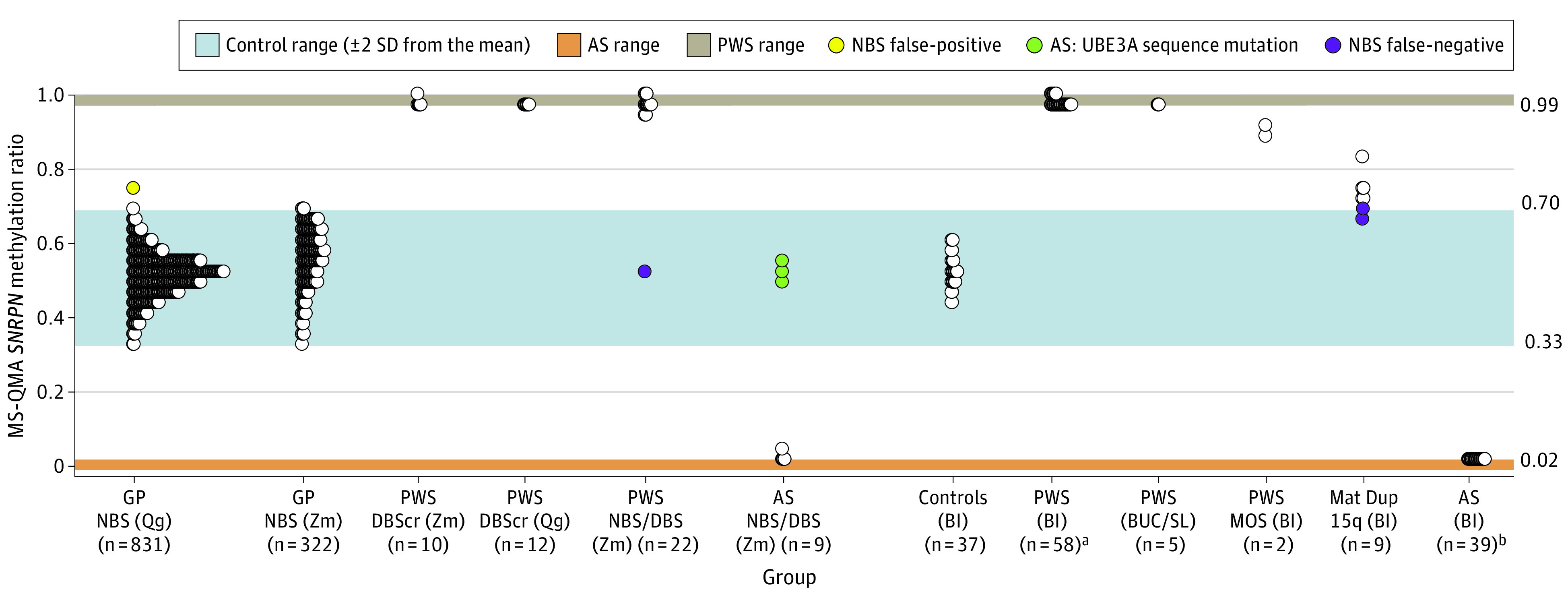
To monitor variability between runs, each 96-well plate had the following controls: (1) a dried blood spot sample from the same Prader-Willi syndrome (PWS) control (denoted as PWS DBScr) and Angelman syndrome (AS) and PWS–spiked DNA samples. NBS indicates newborn blood spots from the general population, showing comparison between Qiagen’s (Qg) and Zymo’s (Zm) bisulfite conversion systems. Of the 22 PWS NBS/DBS samples, 10 were from NBS, and 12 were from DBS made at time of recruitment. Of the 10 AS NBS/DBS samples, 3 were from NBS, and 7 were from DBS made at time of recruitment. Blood (BL), buccal epithelial cell (BUC), and saliva (SL) DNA had high quality from standard diagnostic testing for chromosome 15 imprinting disorders. Mat indicates maternal; MOS, mosaic PWS confirmed through standard diagnostic testing.
aSamples from a total of 44 individuals with AS, with 2 not overlapping between the AS (BL) and AS (NBS/DBS) groups.
bSamples from a total of 72 individuals with PWS, with 7 not overlapping between the PWS (BL) and PWS (NBS/DBS) groups.
