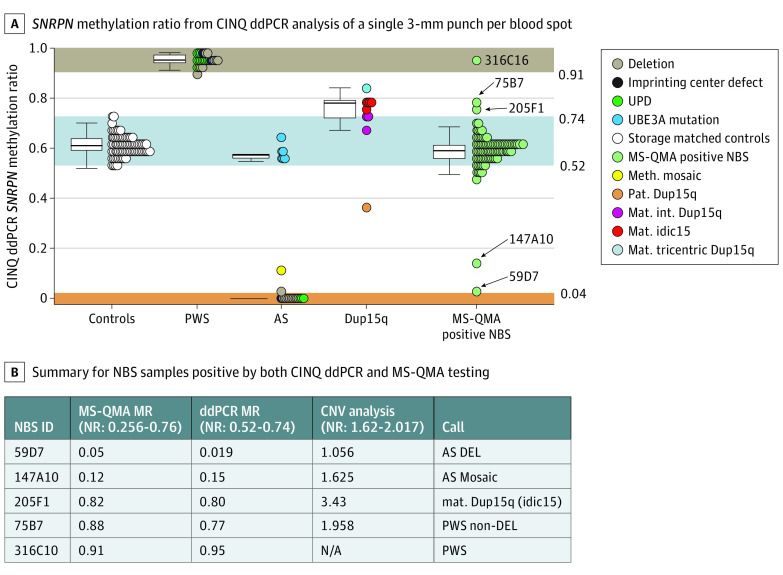
Figure 3. Confirmatory SNRPN Promoter Methylation Testing Using Competitive Priming Initiated Nested Quantification Using Droplet Digital PCR (CINQ ddPCR) on Newborn Blood Spot (NBS) Samples Tested Positive by First-Tier MS-QMA Screening.
A, SNRPN methylation ratio from CINQ ddPCR analysis of a single 3 mm punch per blood spot, including 20 storage-matched control NBS samples (from the general population with MS-QMA methylation ratio of 0.5), dried blood spot samples from 25 patients with PWS, 22 patients with AS, and 11 patients with Dup15q syndrome identified as part of standard diagnostic testing, as well as 92 NBS samples that tested positive by MS-QMA analysis, as part of first-tier testing. NBS IDs are included for the 5 samples confirmed to have abnormal SNRPN promoter methylation. B, Summary of NBS samples that were positive by both CINQ ddPCR and MS-QMA testing from Figure 2 and results on the SNRPN copy number variation (CNV) analysis from Figure 4, with calls made based on neurotypical control reference ranges (NR) and methylation ratio reference ranges in panel A from DBS samples from patients with diagnosis confirmed by standard of care diagnostic testing. N/A indicates value not available.