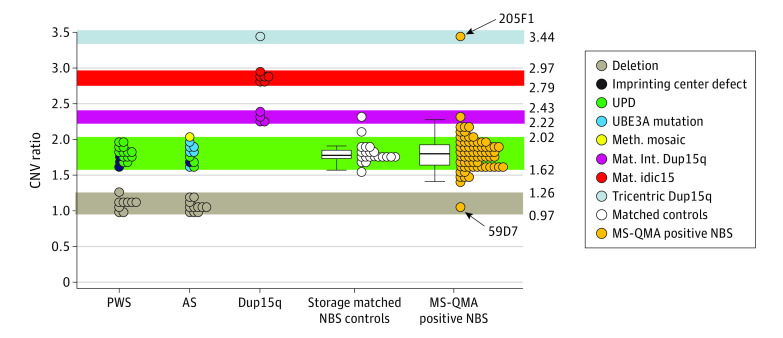
Figure 4. Confirmatory SNRPN Copy Number Variation (CNV) Testing Using Real-Time Polymerase Chain Reaction (PCR) Relative Standard Curve Method on Newborn Blood Spot (NBS) Samples That Were Positive by First-Tier MS-QMA Testing.
Of the 92 NBS samples shortlisted from first-tier testing that were tested by CINQ ddPCR, sufficient DNA was available for only 69 NBS samples for CNV confirmatory testing. SNRPN CNV analysis was performed using real-time PCR relative standard curve method of a single 3 mm punch per blood spot, including dried blood spot (DBS) samples from 25 patients with PWS, 22 patients with AS, and 11 patients with Dup15q syndrome identified as part of standard diagnostic testing, as well as 20 storage-matched control NBS samples (from the general population with MS-QMA methylation ratio of 0.5) and 69 NBS samples that were positive by quantitative MS-QMA analysis. For all DBS samples used to establish CNV reference ranges, the copy number of the PWS/AS imprinted region was confirmed in diagnostic settings using chromosomal microarray. The 1-copy reference range (highlighted in brown) between 0.97 and 1.26 CNV ratio units was established using minimum and maximum values from 20 deletion cases (PWS and AS deletion groups collapsed). The 2-copy reference range (highlighted in green) between 1.62 and 2.02 CNV ratio was established using minimum and maximum values from 27 nondeletion cases (PWS and AS nondeletion groups collapsed). The 4- and 5-copy reference ranges (highlighted in purple and gray) were established using minimum and maximum values from DBS samples from 4 interstitial 15q duplication and 6 isodicentric 15q patients. Although the 7-copy reference range could not be established because there was only 1 patient with tricentric 15q, the reference value for that 1 DBS sample has been included (highlighted in light blue). Black arrows point to 2 samples with the highest and lowest values overlapping with tricentric 15q and deletion CNV ranges that were reflexed to LC-WGS confirmatory testing.