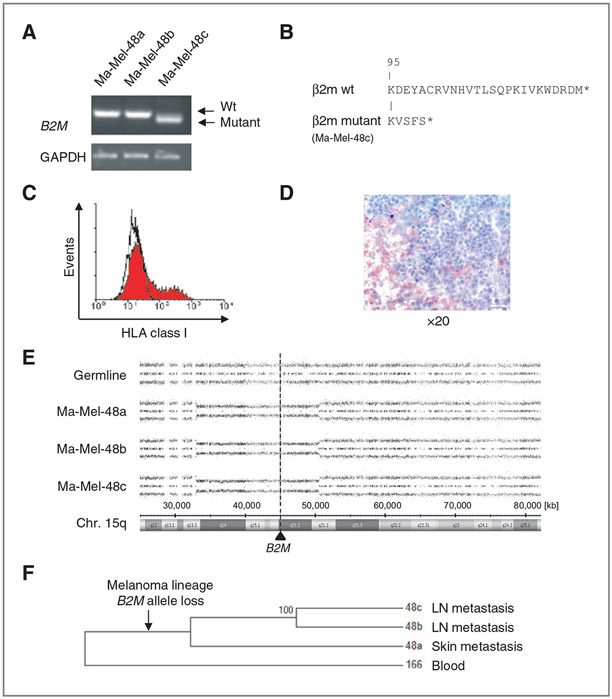
Figure 5.
β2m deficiency of Ma-Mel-48c cells is caused by B2M gene mutation subsequent to B2M allele loss. A, semiquantitative analysis of B2M mRNA expression by RT-PCR. Ma-Mel-48c cells express a shortened B2M mRNA, a PCR product in the size of wild-type (wt) mRNA was only detectable in Ma-Mel-48a and Ma-Mel-48b cells. B, comparison of the C-terminal amino acid sequence (aa) of mutant β2m, as expressed in Ma-Mel-48c cells, and wt β2m. Capital letters indicate the aa sequence. The first 95 aa are identical in wt and mutant proteins. Stars indicate stop codons. C, Ma-Mel-48c cells were transfected with a B2M expression plasmid. Expression of HLA class I antigens on transient transfectants was determined by flow cytometry. D, analysis of β2m expression in metastatic lesion Ma-Mel-48c by IHC. Tumor cells were negative for β2m and did not stain red. E, SNP results given as allelic distribution of chromosome 15q are shown for DNA obtained from autologous PBMC and melanoma cells. All Ma-Mel-48 cell lines show loss of one chromosomal allele in the region 15q13.3 to 15q21.2 (Chr.15: 33,045,756-50,579,508; hg19). The location of B2M at 15q21.1 (Chr.15: 45,003,675-45,011,075) is shown by the dashed line. F, maximum parsimony tree showing the phylogenetic relationship of the melanoma cell lines and the blood sample (166) used as outgroup. Within the patient, a melanoma lineage leading to the studied cell lines evolved and was genetically divergent from the blood sample. A B2M loss evolved on this lineage. Then, a lineage leading to Ma-Mel-48a diverged from the melanoma ancestor and cumulated genotypic differences specific to this lineage. Later the lineages of Ma-Mel-48b and Ma-Mel-48c diverged and cumulated their specific genotypic differences. Hundred percent of the bootstrap replicates showed this grouping.