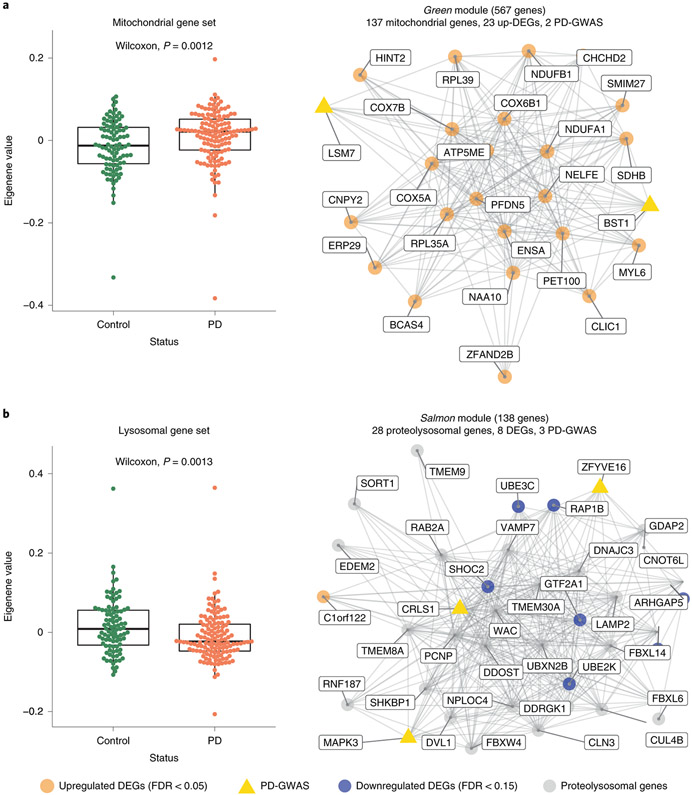
Figure 3. Co-expression networks in monocytes capture PD-specific processes.
(A) Eigengene analysis of all genes in the “mitochondrial” GO category (n = 1302) between PD and controls (Left panel; Two sided Wilcoxon rank-signed test, P-value = 0.0012). Example of a module (green) enriched for PD heritability, mitochondrial genes, and upregulated DEGs (Right panel). Edges represent co-expression connectivity. Nodes in orange are upregulated DEGs at FDR < 0.05; yellow triangles are genes in PD GWAS loci. (B) Eigengene analysis of all genes in the “lysosome” GO category (n = 526) between PD and controls (Two sided Wilcoxon rank sum test, P-value = 0.0013) (Left panel). Example of a module (salmon) enriched for PD heritability, proteo-lysosomal genes, and downregulated DEGs. Nodes in orange are upregulated DEGs at FDR < 0.05; grey are selected proteo-lysosomal genes; and yellow triangles are genes in PD GWAS loci (Right panel). Boxplots: the line represents the median. The boxes extend from the 25th - 75th percentile and the lines extend 1.5 times the interquartile range.